1GPD
 
 | | STUDIES OF ASYMMETRY IN THE THREE-DIMENSIONAL STRUCTURE OF LOBSTER D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Moras, D, Olsen, K.W, Sabesan, M.N, Buehner, M, Ford, G.C, Rossmann, M.G. | | Deposit date: | 1975-07-01 | | Release date: | 1977-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase.
J.Biol.Chem., 250, 1975
|
|
2A3V
 
 | | Structural basis for broad DNA-specificity in integron recombination | | Descriptor: | DNA (31-MER), DNA (34-MER), site-specific recombinase IntI4 | | Authors: | MacDonald, D, Demarre, G, Bouvier, M, Mazel, D, Gopaul, D.N. | | Deposit date: | 2005-06-27 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for broad DNA-specificity in integron recombination.
Nature, 440, 2006
|
|
4W63
 
 | | TORPEDO CALIFORNICA ACETYLCHOLINESTERASE IN COMPLEX WITH A TACRINE-BENZOFURAN HYBRID INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pesaresi, A, Samez, S, Lamba, D. | | Deposit date: | 2014-08-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Tacrine-Benzofuran Hybrids as Potent Multitarget-Directed Ligands for the Treatment of Alzheimer's Disease: Design, Synthesis, Biological Evaluation, and X-ray Crystallography.
J.Med.Chem., 59, 2016
|
|
4V0C
 
 | |
5J41
 
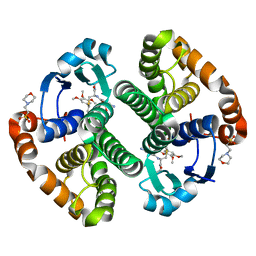 | | Glutathione S-transferase bound with hydrolyzed Piperlongumine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(3,4,5-trimethoxyphenyl)propanoic acid, GLUTATHIONE, ... | | Authors: | Harshbarger, W, Gondi, S, Ficarro, S, Hunter, J, Udayakumar, D, Gurbani, D, Marto, J, Westover, K. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19035351 Å) | | Cite: | Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine.
J. Biol. Chem., 292, 2017
|
|
4V02
 
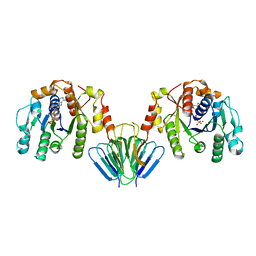 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
1HLW
 
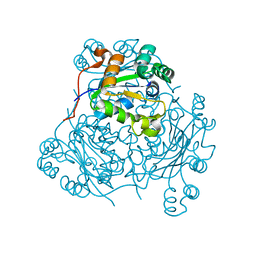 | | STRUCTURE OF THE H122A MUTANT OF THE NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Admiraal, S.J, Meyer, P, Schneider, B, Deville-Bonne, D, Janin, J, Herschlag, D. | | Deposit date: | 2000-12-04 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical rescue of phosphoryl transfer in a cavity mutant: a cautionary tale for site-directed mutagenesis.
Biochemistry, 40, 2001
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
4UXZ
 
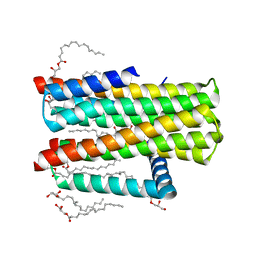 | | Structure of delta7-DgkA-syn in 7.9 MAG to 2.18 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, ACETATE ION, ... | | Authors: | Li, D, Howe, N, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
3BPX
 
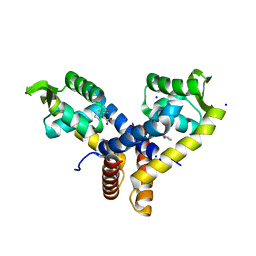 | | Crystal Structure of MarR | | Descriptor: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
7A2B
 
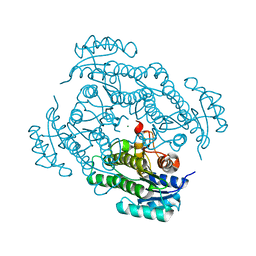 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q207D | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Bischoff, D, Hermann, J, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-08-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
2AK5
 
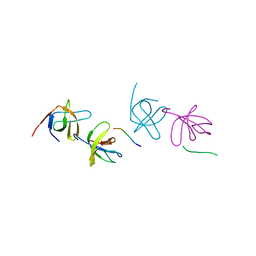 | | beta PIX-SH3 complexed with a Cbl-b peptide | | Descriptor: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | Authors: | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
2ATI
 
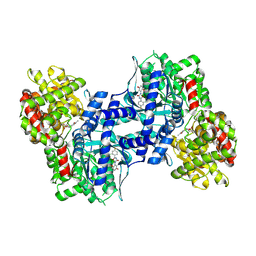 | | Glycogen Phosphorylase Inhibitors | | Descriptor: | Glycogen phosphorylase, liver form, N-(2-CHLORO-4-FLUOROBENZOYL)-N'-(5-HYDROXY-2-METHOXYPHENYL)UREA, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.J, Herling, A.W, Oikonomakos, N.G, Schmoll, D, Sarubbi, E, von Roedern, E, Schoenafinger, K, Defossa, E. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes.
J.Med.Chem., 48, 2005
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
1QMA
 
 | | Nuclear Transport Factor 2 (NTF2) W7A mutant | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Bayliss, R, Ribbeck, K, Akin, D, Kent, H.M, Feldherr, C.M, Gorlich, D, Stewart, M.J. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction Betweeen Ntf2 and Xfxfg-Containing Nucleoporins is Required to Mediate Nuclear Import of Ran-Gdp
J.Mol.Biol., 293, 1999
|
|
1QW9
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | Descriptor: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
3CBP
 
 | | Set7/9-ER-Sinefungin complex | | Descriptor: | BETA-MERCAPTOETHANOL, Estrogen receptor, GLYCEROL, ... | | Authors: | Cheng, X, Jia, D. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase.
Mol.Cell, 30, 2008
|
|
5WYH
 
 | | Crystal structure of RidL(1-200) complexed with VPS29 | | Descriptor: | GLYCEROL, Interaptin, Vacuolar protein sorting-associated protein 29 | | Authors: | Yao, J, Sun, Q, Jia, D. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of inhibition of retromer transport by the bacterial effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1QW8
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1R4T
 
 | | Solution structure of exoenzyme S | | Descriptor: | exoenzyme S | | Authors: | Langdon, G.M, Leitner, D, Labudde, D, Kuhne, R, Schmieder, P, Aktories, K, Oschkinat, H.O, Schmidt, G. | | Deposit date: | 2003-10-08 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal GTPase activating domain of Pseudomonas aeruginosa exoenzyme S
To be Published
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1H6B
 
 | |
5J36
 
 | | Crystal structure of 60-mer BFDV Capsid Protein | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
