6NUG
 
 | | hGRNA4-28_3s | | Descriptor: | Granulin-4 | | Authors: | Dastpeyman, M, Bansal, P, Wilson, D, Loukas, A, Smout, M, Daly, N. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | hGRN4-28_3s
to be published
|
|
8GQ4
 
 | |
2YBT
 
 | | Crystal structure of human acidic chitinase in complex with bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), ACIDIC MAMMALIAN CHITINASE, GLYCEROL | | Authors: | Sutherland, T.E, Andersen, O.A, Betou, M, Eggleston, I.M, Maizels, R.M, van Aalten, D, Allen, J.E. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor.
Chem. Biol., 18, 2011
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
4WH9
 
 | | Structure of the CDC25B Phosphatase Catalytic Domain with Bound Inhibitor | | Descriptor: | 2-[(2-cyano-3-fluoro-5-hydroxyphenyl)sulfanyl]ethanesulfonic acid, GLYCEROL, M-phase inducer phosphatase 2, ... | | Authors: | Lund, G.L, Dudkin, S, Borkin, D, Ni, W, Grembecka, J, Cierpicki, T. | | Deposit date: | 2014-09-20 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction.
Acs Chem.Biol., 10, 2015
|
|
1DAW
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF PROTEIN KINASE CK2 (ALPHA-SUBUNIT) AND MG-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN KINASE CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.G, Schomburg, D. | | Deposit date: | 1999-11-01 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GTP plus water mimic ATP in the active site of protein kinase CK2.
Nat.Struct.Biol., 6, 1999
|
|
2YOC
 
 | | Crystal structure of PulA from Klebsiella oxytoca | | Descriptor: | CALCIUM ION, PULLULANASE, SULFATE ION | | Authors: | Francetic, O, Mechaly, A.E, Tello-Manigne, D, Buschiazzo, A, Bernarde, C, Nadeau, N, Pugsley, A.P, Alzari, P.M. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-06 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of Pullulanase Membrane Binding and Secretion Revealed by X-Ray Crystallography, Molecular Dynamics and Biochemical Analysis
Structure, 24, 2016
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
8GKO
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | 1,2-ETHANEDIOL, Alkaline protease 1, FORMYL GROUP, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
8G4L
 
 | | Cryo-EM structure of the human cardiac myosin filament | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | Dutta, D, Nguyen, V, Padron, R, Craig, R. | | Deposit date: | 2023-02-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-EM structure of the human cardiac myosin filament.
Nature, 623, 2023
|
|
8GKQ
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | 1,2-ETHANEDIOL, Alkaline protease 1, CALCIUM ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
8GKP
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-03-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
6OPI
 
 | | phosphorylated ERK2 with SCH-CPD336 | | Descriptor: | (3R)-N-[3-(2-cyclopropylpyridin-4-yl)-1H-indazol-5-yl]-3-(methoxymethyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1DJ9
 
 | | CRYSTAL STRUCTURE OF 8-AMINO-7-OXONANOATE SYNTHASE (OR 7-KETO-8AMINIPELARGONATE OR KAPA SYNTHASE) COMPLEXED WITH PLP AND THE PRODUCT 8(S)-AMINO-7-OXONANONOATE (OR KAPA). THE ENZYME OF BIOTIN BIOSYNTHETIC PATHWAY. | | Descriptor: | 8-AMINO-7-OXONONANOATE SYNTHASE, MAGNESIUM ION, N-[7-KETO-8-AMINOPELARGONIC ACID]-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
4WL3
 
 | |
4WIH
 
 | | Crystal structure of cAMP-dependent Protein Kinase A from Cricetulus griseus | | Descriptor: | cAMP Dependent Protein Kinase Inhibitor PKI-tide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Kudlinzki, D, Linhard, V.L, Saxena, K, Dreyer, M, Schwalbe, H. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | High-resolution crystal structure of cAMP-dependent protein kinase from Cricetulus griseus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6O6R
 
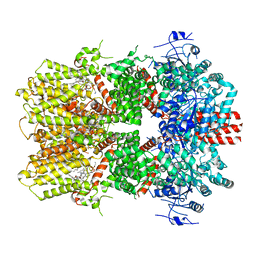 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, AMTB-bound state | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, CHOLESTEROL HEMISUCCINATE, N-(3-aminopropyl)-2-[(3-methylphenyl)methoxy]-N-[(thiophen-2-yl)methyl]benzamide, ... | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
6UAN
 
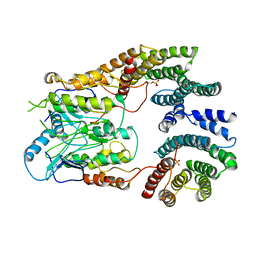 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
4WFA
 
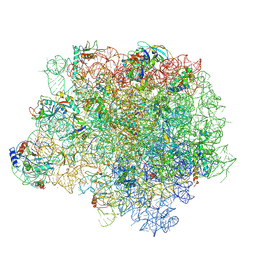 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with linezolid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6O7X
 
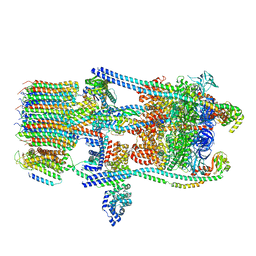 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4WGF
 
 | | YcaC from Pseudomonas aeruginosa with hexane-2,5-diol and covalent acrylamide | | Descriptor: | (2R,5R)-hexane-2,5-diol, CHLORIDE ION, PROPIONAMIDE, ... | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34020686 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
1DDT
 
 | |
6OAN
 
 | | Structure of DBP in complex with human neutralizing antibody 053054 | | Descriptor: | Antibody 053054 single chain variable fragment, Duffy binding surface protein region II, SULFATE ION | | Authors: | Urusova, D, Tolia, N.H. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP.
Nat Microbiol, 4, 2019
|
|
4GL5
 
 | |
4WO2
 
 | |
