8C0E
 
 | | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase Wzz form a complex in vivo and in vitro | | Descriptor: | ECA polysaccharide chain length modulation protein | | Authors: | Weckener, M, Woodward, L.S, Clarke, B.R, Liu, H, Ward, P.N, Le Bas, A, Bhella, D, Whitfield, C, Naismith, J.H. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase, Wzz, from enterobacterial common antigen biosynthesis form a complex.
Open Biology, 13, 2023
|
|
6UPR
 
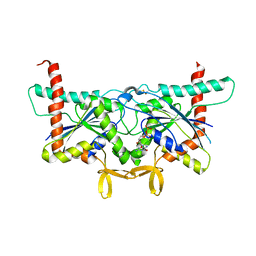 | | Crystal Structure of GTPase Domain of Human Septin 2 / Septin 8 Heterocomplex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Leonardo, D.A, Pereira, H.M, Brandao-Neto, J, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2019-10-18 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Molecular Recognition at Septin Interfaces: The Switches Hold the Key.
J.Mol.Biol., 432, 2020
|
|
5CLW
 
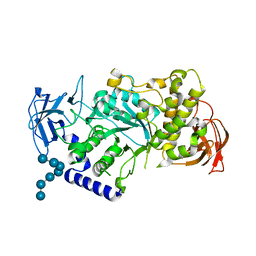 | | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose | | Descriptor: | 1,4-alpha-glucan-branching enzyme, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krojer, T, Froese, D.S, Goubin, S, Strain-Damerell, C, Mahajan, P, Burgess-Brown, N, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human glycogen branching enzyme (GBE1) in complex with maltoheptaose
To be published
|
|
8C0C
 
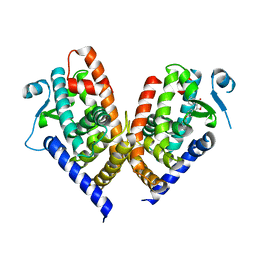 | | X-ray crystal structure of PPAR gamma ligand binding domain in complex with CZ46 | | Descriptor: | (2~{R})-2-[4-(naphthalen-1-ylmethoxy)phenyl]-4-oxidanyl-3-phenyl-2~{H}-furan-5-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R, Pochetti, G, Villa, S, Meneghetti, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biological Screening and Crystallographic Studies of Hydroxy gamma-Lactone Derivatives to Investigate PPAR gamma Phosphorylation Inhibition.
Biomolecules, 13, 2023
|
|
6F5V
 
 | | Crystal structure of the prephenate aminotransferase from Arabidopsis thaliana | | Descriptor: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Robin, A, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-03 | | Release date: | 2019-03-13 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
5FJ4
 
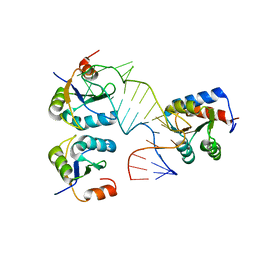 | |
7AZP
 
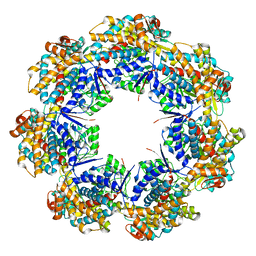 | |
6UQW
 
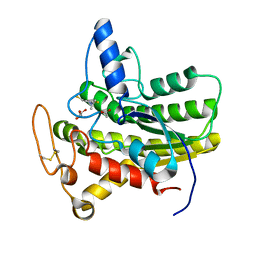 | |
7MZY
 
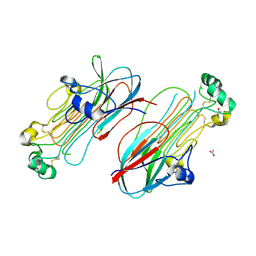 | | Anaplastic lymphoma kinase (ALK) extracellular fragment of ligand binding region 673-986 | | Descriptor: | ACETATE ION, ALK tyrosine kinase receptor | | Authors: | Reshetnyak, A.V, Sowaileh, M, Miller, D.J, Rossi, P, Myasnikov, A.G, Kalodimos, C.G. | | Deposit date: | 2021-05-24 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism for the activation of the anaplastic lymphoma kinase receptor.
Nature, 600, 2021
|
|
6UR1
 
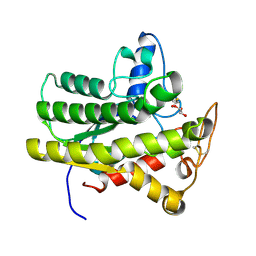 | |
7ATI
 
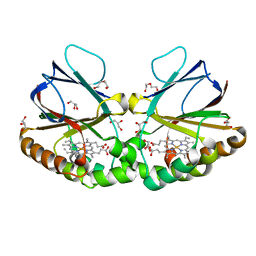 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
6P3O
 
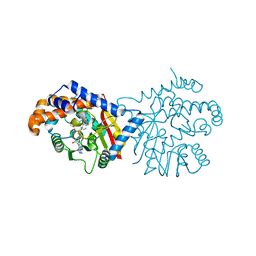 | | Tetrahydroprotoberberine N-methyltransferase in complex with (S)-cis-N-methylstylopine and S-adenosylhomocysteine | | Descriptor: | (5S,12bS)-5-methyl-6,7,12b,13-tetrahydro-2H,4H,10H-[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquinolino[3,2-a]isoquinolin-5-ium, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
6RKC
 
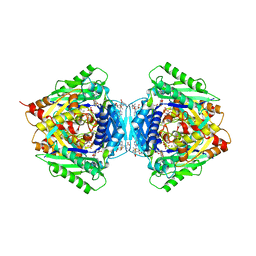 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6XN8
 
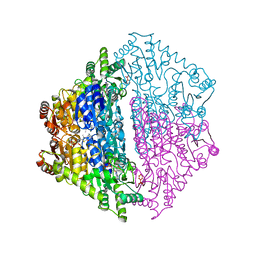 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
8C0Z
 
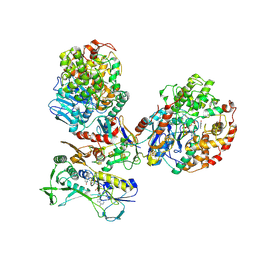 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
6F12
 
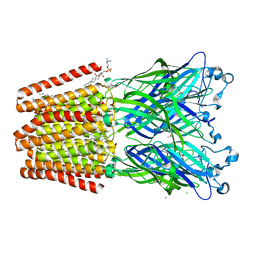 | | GLIC mutant E181A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
7ASB
 
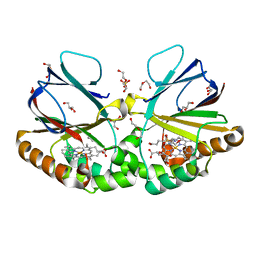 | | Crystal structure of dimeric chlorite dismutase variant Q74E (CCld Q74E) from Cyanothece sp. PCC7425 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chlorite dismutase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-27 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
6UQZ
 
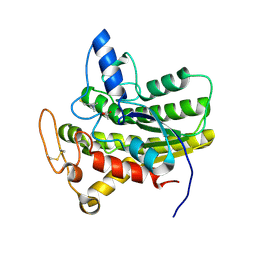 | |
8EIU
 
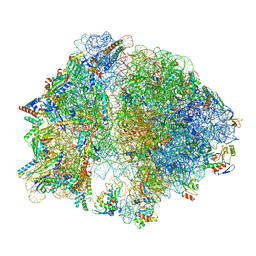 | | E. coli 70S ribosome with A-loop mutations U2554C and U2555C | | Descriptor: | 16S rRNA, 23S rRNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Nissley, A.J, Penev, P.I, Watson, Z.L, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Rare ribosomal RNA sequences from archaea stabilize the bacterial ribosome.
Nucleic Acids Res., 51, 2023
|
|
6B19
 
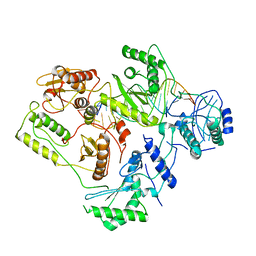 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | Authors: | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | Deposit date: | 2017-09-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
4XGW
 
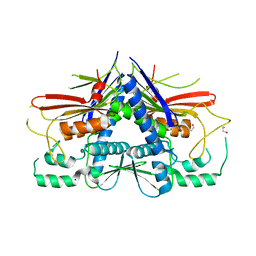 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase, E169K mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
8T0T
 
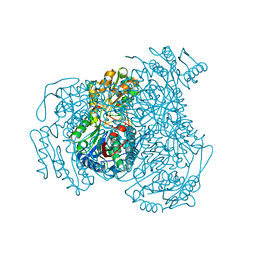 | | Structure of Compound 4 bound to human ALDH1A1 | | Descriptor: | 1-(4-{6-fluoro-3-[4-(methanesulfonyl)piperazine-1-carbonyl]quinolin-4-yl}phenyl)cyclopropane-1-carbonitrile, Aldehyde dehydrogenase 1A1, CHLORIDE ION, ... | | Authors: | Hurley, T.D. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of substituted benzimidazoles as inhibitors of human aldehyde dehydrogenase 1A isoenzymes.
Chem.Biol.Interact., 391, 2024
|
|
7Z19
 
 | | E. coli C-P lyase bound to a single PhnK ABC domain | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7ZEH
 
 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7Guanine | | Descriptor: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, D-ribulose, ... | | Authors: | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
6UVQ
 
 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
