6U9G
 
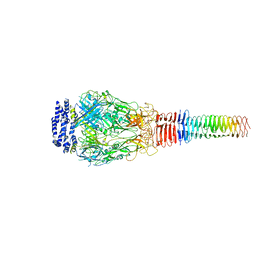 | | Structure of Francisella PdpA-VgrG Complex, half-lidded | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y, Zhou, Z.H, Horwitz, M.A. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
6OKT
 
 | |
7QU9
 
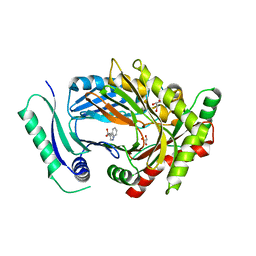 | | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii. | | Descriptor: | Anthranilate synthase component I family protein, GLYCEROL, TRYPTOPHAN | | Authors: | Rooms, L.D, Race, P.R, Back, C.B, Burton, N.B, Willis, C.L, Stach, J.E.M, Duke, P.W, Hawkins, C. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aminodeoxychorismate synthase component 1 (PabB) from Bacillus subtilis spizizenii.
To Be Published
|
|
1D17
 
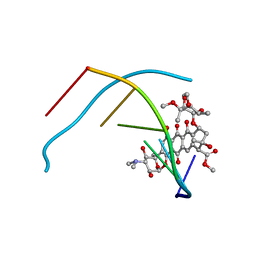 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
3IED
 
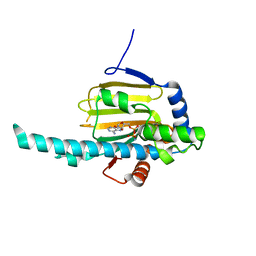 | | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, Heat shock protein | | Authors: | Pizarro, J.C, Wernimont, A.K, Lew, J, Hutchinson, A, Artz, J.D, Amaya, M.F, Plotnikova, O, Vedadi, M, Kozieradzki, I, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN
TO BE PUBLISHED
|
|
6XS4
 
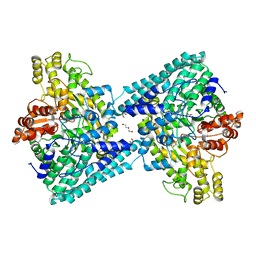 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
8FU2
 
 | |
8FU5
 
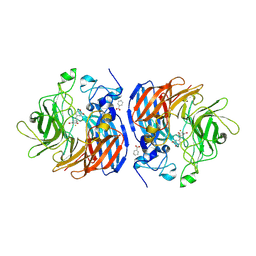 | |
3IJF
 
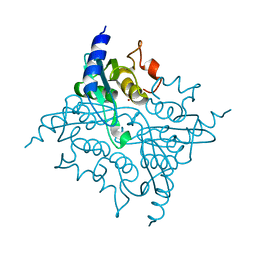 | |
5IAR
 
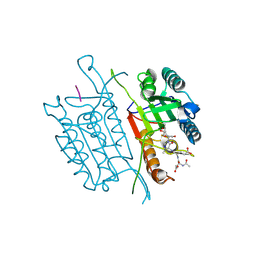 | | Caspase 3 V266W | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASK, Caspase-3, ... | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TYM
 
 | | KEAP1 Kelch domain in complex with Compound 9 | | Descriptor: | (3S)-3-[2-(benzenecarbonyl)-5-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl]-3-(1-ethyl-4-methyl-1H-benzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.422 Å) | | Cite: | Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6XWP
 
 | |
8GAP
 
 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
3OV5
 
 | |
5IMU
 
 | | A fragment of conserved hypothetical protein Rv3899c (residues 184-410) from Mycobacterium tuberculosis | | Descriptor: | POTASSIUM ION, Tat (Twin-arginine translocation) pathway signal sequence containing protein | | Authors: | Li, D.F, Gao, Y.R, Liu, Y.Y, Bi, L.J. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv3899c(184-410), a hypothetical protein from Mycobacterium tuberculosis
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6XXZ
 
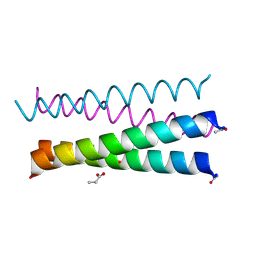 | |
6I00
 
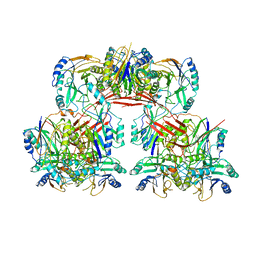 | |
8GR2
 
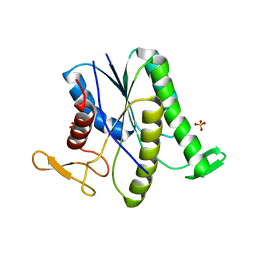 | |
6L34
 
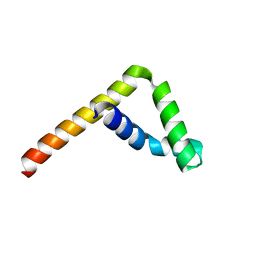 | |
3F2Z
 
 | | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B | | Descriptor: | uncharacterized protein BF3579 | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, Foote, E.L, Ciccosanti, C, Lee, D, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B
To be Published
|
|
4QAO
 
 | | Lysine-ligated cytochrome c with F82H | | Descriptor: | Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Amacher, J.F, Zhu, M.Q, Zhong, F, Pletneva, E.K, Madden, D.R. | | Deposit date: | 2014-05-05 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Understanding PDZ Affinity and Selectivity: All Residues Considered
Thesis, 2014
|
|
8A5Q
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
6TYP
 
 | | KEAP1 Kelch domain in complex with Compound 2 | | Descriptor: | (3S)-3-[2-(benzenecarbonyl)-1,2,3,4-tetrahydroisoquinolin-7-yl]-3-(1-ethyl-4-methyl-1H-benzotriazol-5-yl)propanoic acid, FORMIC ACID, Kelch-like ECH-associated protein 1 | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4QEW
 
 | | Crystal structure of BRD2(BD2) mutant with ligand ET bound (METHYL (2R)- 2-[(4S)-6-(4-CHLOROPHENYL)-8-METHOXY-1-METHYL-4H-[1,2,4]TRIAZOLO[4,3-A][1, 4]BENZODIAZEPIN-4-YL]BUTANOATE) | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical biology. A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes.
Science, 346, 2014
|
|
6UID
 
 | |
