1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
3T67
 
 | |
2EI0
 
 | | Anaerobic Crystal Structure Analysis of 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexed with 3,4-dihydroxybiphenyl | | Descriptor: | 1,1'-BIPHENYL-3,4-DIOL, 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G5S
 
 | | CRYSTAL STRUCTURE OF HUMAN CYCLIN DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH THE INHIBITOR H717 | | Descriptor: | 2-[TRANS-(4-AMINOCYCLOHEXYL)AMINO]-6-(BENZYL-AMINO)-9-CYCLOPENTYLPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Dreyer, M.K, Borcherding, D.R, Dumont, J.A, Peet, N.P, Tsay, J.T, Wright, P.S, Bitonti, A.J, Shen, J, Kim, S.-H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of human cyclin-dependent kinase 2 in complex with the adenine-derived inhibitor H717.
J.Med.Chem., 44, 2001
|
|
1G5Z
 
 | | CRYSTAL STRUCTURE OF LYME DISEASE ANTIGEN OUTER SURFACE PROTEIN C (OSPC) FROM BORRELIA BURGDORFERI STRAIN N40 | | Descriptor: | OUTER SURFACE PROTEIN C | | Authors: | Eicken, C, Sharma, V, Klabunde, T, Owens, R.T, Pikas, D.S, Hook, M, Sacchettini, J.C. | | Deposit date: | 2000-11-02 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi.
J.Biol.Chem., 276, 2001
|
|
6DAU
 
 | | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | GLYCEROL, Spermidine N1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Beahan, A, Kulyavtsev, P, Tan, L, Tran, D, Kuhn, M.L, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To be Published
|
|
2PFE
 
 | | Crystal Structure of Thermobifida fusca Protease A (TFPA) | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, Alkaline serine protease, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-04-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.436 Å) | | Cite: | Mesophile versus Thermophile: Insights Into the Structural Mechanisms of Kinetic Stability
J.Mol.Biol., 370, 2007
|
|
2PPO
 
 | |
2PTF
 
 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
2PP8
 
 | | Formate bound to oxidized wild type AfNiR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Tocheva, E.I, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2007-04-28 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved active site residues limit inhibition of a copper-containing nitrite reductase by small molecules.
Biochemistry, 47, 2008
|
|
6DTK
 
 | | Heterodimers of FALS mutant SOD enzyme | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase C111S/D83S-C111S HETERODIMER, ... | | Authors: | Streltsov, V.A, Nuttall, S.D, Ganio, K.E, Roberts, B. | | Deposit date: | 2018-06-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of heterodimers of FALS mutant SOD enzyme
To Be Published
|
|
2PWZ
 
 | |
1GA0
 
 | | STRUCTURE OF THE E. CLOACAE GC1 BETA-LACTAMASE WITH A CEPHALOSPORIN SULFONE INHIBITOR | | Descriptor: | 3-(4-CARBAMOYL-1-CARBOXY-2-METHYLSULFONYL-BUTA-1,3-DIENYLAMINO)-INDOLIZINE-2-CARBOXYLIC ACID, BETA-LACTAMASE, GLYCEROL, ... | | Authors: | Crichlow, G.V, Nukaga, M, Buynak, J.D, Knox, J.R. | | Deposit date: | 2000-11-28 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of class C beta-lactamases: structure of a reaction intermediate with a cephem sulfone.
Biochemistry, 40, 2001
|
|
3T3X
 
 | |
2PLL
 
 | | Crystal structure of perdeuterated human arginase I | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, arginase-1 | | Authors: | Di Costanzo, L, Moulin, M, Haertlein, M, Meilleur, F, Christianson, D.W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification, assay, and crystal structure of perdeuterated human arginase I
Arch.Biochem.Biophys., 465, 2007
|
|
8RB5
 
 | |
7M2Y
 
 | | Closed conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Z
 
 | | Monomeric single-particle reconstruction of the Yeast gamma-TuSC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component SPC97, Spindle pole body component SPC98, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
8RB3
 
 | |
8RB7
 
 | |
1G1H
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A BIS-PHOSPHORYLATED PEPTIDE (ETD(PTR)(PTR)RKGGKGLL) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | BI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
7M2X
 
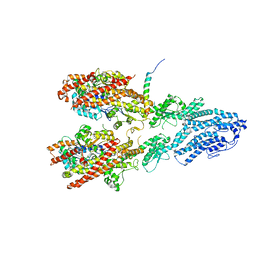 | | Open conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
1G2N
 
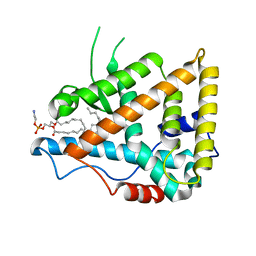 | | CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE ULTRASPIRACLE PROTEIN USP, THE ORTHOLOG OF RXRS IN INSECTS | | Descriptor: | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ULTRASPIRACLE PROTEIN | | Authors: | Billas, I.M.L, Moulinier, L, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects.
J.Biol.Chem., 276, 2001
|
|
