5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
8SG0
 
 | | Crystal Structure of GDP-manose 3,5 epimerase de Myrciaria dubia in complex with substrate, product and NAD | | Descriptor: | GDP-mannose 3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-GALACTOSE, ... | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
7SGC
 
 | |
2G1N
 
 | | Ketopiperazine-based renin inhibitors: Optimization of the "C" ring | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-{2-[6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2,2-DIMETHYL-3-OXO-2,3-DIHYDRO-4H-1,4-BENZOTHIAZIN-4-YL]ETHYL}ACETAMIDE, Renin | | Authors: | Holsworth, D.D. | | Deposit date: | 2006-02-14 | | Release date: | 2006-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ketopiperazine-based renin inhibitors: Optimization of the "C" ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
2GH6
 
 | | Crystal structure of a HDAC-like protein with 9,9,9-trifluoro-8-oxo-N-phenylnonan amide bound | | Descriptor: | 9,9,9-TRIFLUORO-8-OXO-N-PHENYLNONANAMIDE, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Riester, D, Wegener, D, Schwienhorst, A, Ficner, R. | | Deposit date: | 2006-03-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Complex structure of a bacterial class 2 histone deacetylase homologue with a trifluoromethylketone inhibitor.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4FET
 
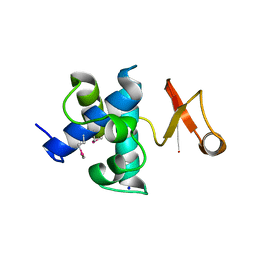 | | Catalytic domain of germination-specific lytic tansglycosylase SleB from Bacillus anthracis | | Descriptor: | SODIUM ION, Spore cortex-lytic enzyme prepeptide | | Authors: | Jing, X, Heffron, J, Popham, D.L, Schubot, F.D. | | Deposit date: | 2012-05-30 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | The catalytic domain of the germination-specific lytic transglycosylase SleB from Bacillus anthracis displays a unique active site topology.
Proteins, 80, 2012
|
|
1HNG
 
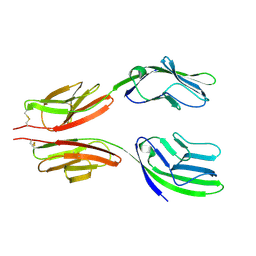 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | Descriptor: | CD2 | | Authors: | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
4WGI
 
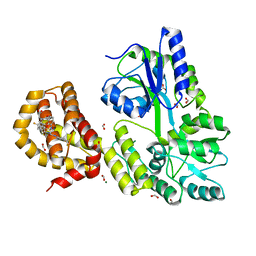 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
3LM1
 
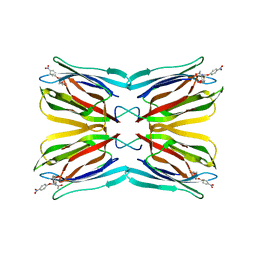 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
4FQV
 
 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FJ0
 
 | | Crystal structure of the ternary complex between a fungal 17beta-hydroxysteroid dehydrogenase (Holo form) and 3,7-dihydroxy flavone | | Descriptor: | 1,2-ETHANEDIOL, 17beta-hydroxysteroid dehydrogenase, 3,7-dihydroxy-2-phenyl-4H-chromen-4-one, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I. | | Deposit date: | 2012-06-11 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inhibition of 17 beta-hydroxysteroid dehydrogenases by phytoestrogens: The case of fungal 17 beta-HSDcl.
J. Steroid Biochem. Mol. Biol., 171, 2017
|
|
4WJT
 
 | | Stationary Phase Survival Protein YuiC from B.subtilis complexed with NAG | | Descriptor: | (2S)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Quay, D.H.X, Cole, A.R, Cryar, A, Thalassinos, K, Williams, M.A, Bhakta, S, Keep, N.H. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure of the stationary phase survival protein YuiC from B.subtilis.
Bmc Struct.Biol., 15, 2015
|
|
2G5N
 
 | |
4FA3
 
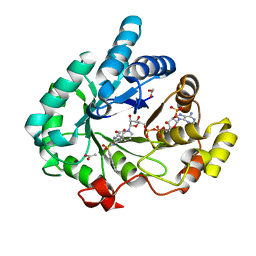 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid (86) | | Descriptor: | (3R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
6DZY
 
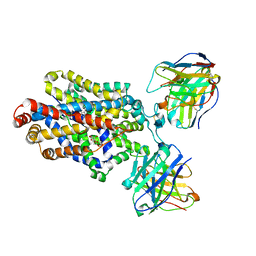 | | Cryo-EM structure of the ts2-active human serotonin transporter in complex with 15B8 Fab and 8B6 ScFv bound to ibogaine | | Descriptor: | (5beta)-12-methoxyibogamine, 15B8 antibody heavy chain, 15B8 antibody light chain, ... | | Authors: | Coleman, J.A, Yang, D, Gouaux, E. | | Deposit date: | 2018-07-05 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport.
Nature, 569, 2019
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
3LH9
 
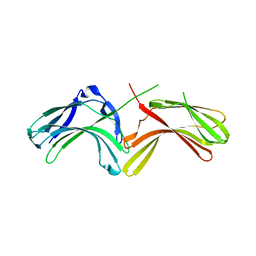 | |
2G43
 
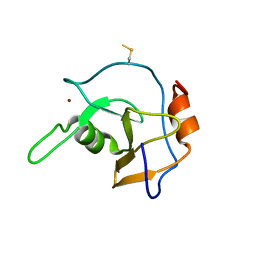 | | Structure of the ZNF UBP domain from deubiquitinating enzyme isopeptidase T (IsoT) | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ZINC ION | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2GBJ
 
 | |
4WOG
 
 | | Crystal Structure of Frutalin from Artocarpus incisa | | Descriptor: | Frutalin | | Authors: | Pereira, H.M, Moreira, A.C.O.M, Vieira Neto, A.E, Moreno, F.B.M.B, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa
To Be Published
|
|
5W27
 
 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
5KJA
 
 | | Synechocystis apocarotenoid oxygenase (ACO) mutant - Trp149Ala | | Descriptor: | Apocarotenoid-15,15'-oxygenase, CHLORIDE ION, FE (II) ION | | Authors: | Sui, X, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-06-18 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Key Residues for Catalytic Function and Metal Coordination in a Carotenoid Cleavage Dioxygenase.
J.Biol.Chem., 291, 2016
|
|
1HNO
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1 | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
6WMQ
 
 | |
7R73
 
 | |
