7SGH
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML124N | | Descriptor: | (S)-N-((S)-1-imino-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)-4-methyl-2-(2-((2,4,6-trifluorophenyl)amino)acetamido)pentanamide, 3C-like proteinase | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
8JT9
 
 | | Human VMAT2 complex with ketanserin | | Descriptor: | 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
6RRH
 
 | | GOLGI ALPHA-MANNOSIDASE II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION | | Authors: | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2019-05-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
7SGJ
 
 | |
3H6O
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(2-fluorobenzyl)-2,4-dimethyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-23 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activator-Bound Structures of Human Pyruvate Kinase M2
to be published
|
|
6F3D
 
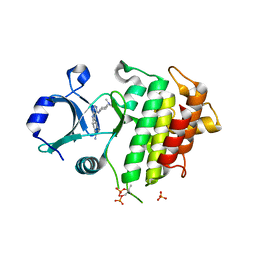 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
8SN3
 
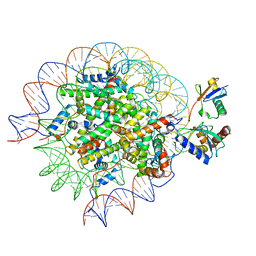 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SMZ
 
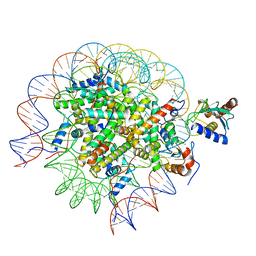 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (Class 4) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8C3X
 
 | |
8SN8
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A) (class 6) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5C9A
 
 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
8SN9
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c with backside ubiquitin (UbcH5c chemically conjugated to histone H2A) (class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8SN0
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 5) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
6TV7
 
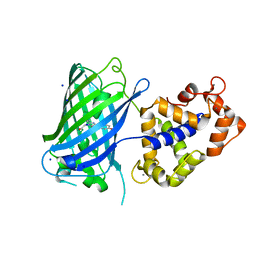 | | Crystal structure of rsGCaMP in the OFF state (illuminated) | | Descriptor: | CALCIUM ION, SODIUM ION, rsGCaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-01-09 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
8KG8
 
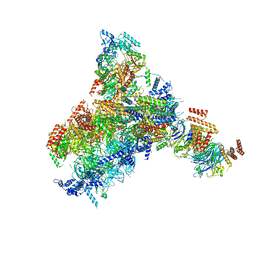 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8SN1
 
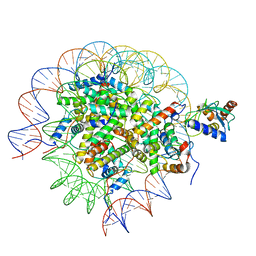 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168 and UbcH5c~Ub (UbcH5c chemically conjugated to histone H2A. No density for Ub.) (class 6) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
3HSC
 
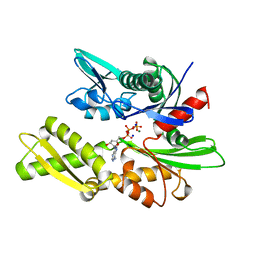 | | THREE-DIMENSIONAL STRUCTURE OF THE ATPASE FRAGMENT OF A 70K HEAT-SHOCK COGNATE PROTEIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 7OKD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Deluca-Flaherty, C.R, Mckay, D.B. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein.
Nature, 346, 1990
|
|
4XMO
 
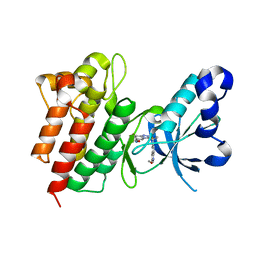 | | Crystal structure of c-Met in complex with (R)-5-(8-fluoro-3-(1-fluoro-1-(3-methoxyquinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1R)-1-fluoro-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-methoxyquinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
7SGI
 
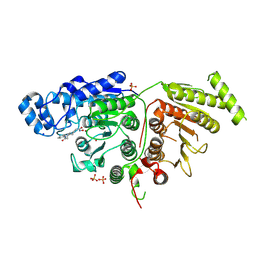 | |
6F19
 
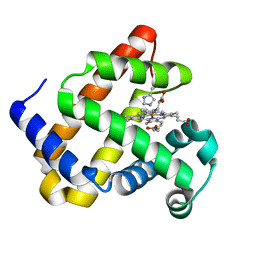 | | Structure of Mb NMH H64V, V68A mutant complex with EDA incubated at room temperature for 5 min | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
8KG9
 
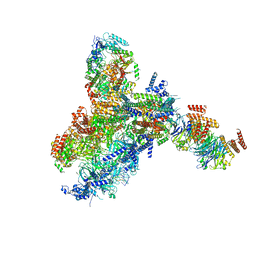 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
7MZ5
 
 | |
7MZ6
 
 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
