1J5L
 
 | | NMR STRUCTURE OF THE ISOLATED BETA_C DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | Deposit date: | 2002-05-16 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
5R5T
 
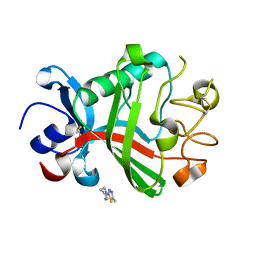 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1251207602 | | Descriptor: | 1-(5-methyl-1,3,4-thiadiazol-2-yl)piperidine, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8W5G
 
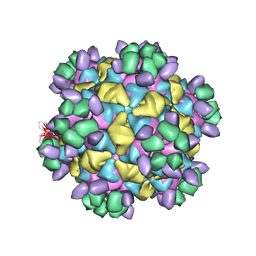 | | Cryo-EM structure of Qb-Ab7 | | Descriptor: | Heavy chain of Ab7, Light chain of Ab7, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-26 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
5I4E
 
 | | Crystal Structure of Human Nonmuscle Myosin 2C motor domain | | Descriptor: | ADP ORTHOVANADATE, MAGNESIUM ION, Myosin-14,Alpha-actinin A | | Authors: | Chinthalapudi, K, Heissler, S.M, Preller, M, Sellers, J.R, Manstein, D.J. | | Deposit date: | 2016-02-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic insights into the active site and allosteric communication pathways in human nonmuscle myosin-2C.
Elife, 6, 2017
|
|
8W5W
 
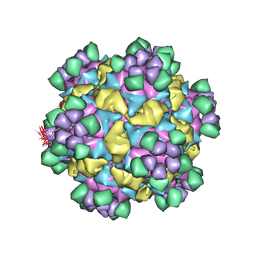 | | Cryo-EM structure of Qb-Ab8 | | Descriptor: | Heavy chain of Ab8, Light chain of Ab8, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
6VPV
 
 | | Trimeric Photosystem I from the High-Light Tolerant Cyanobacteria Cyanobacterium Aponinum | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Dobson, Z, Toporik, H, Vaughn, N, Lin, S, Williams, D, Fromme, P, Mazor, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of photosystem I from a high-light tolerant Cyanobacteria.
Elife, 10, 2021
|
|
6Q84
 
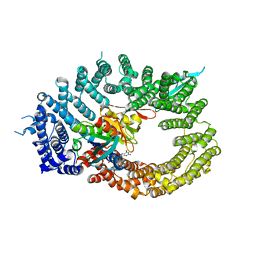 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
8W5R
 
 | | Cryo-EM structure of Qb-Ab53 | | Descriptor: | Heavy chain of Ab53, Light chain of Ab53, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
7F17
 
 | | Crystal Structure of acid phosphatase | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
6QAG
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-1,2,3-triazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
8W5P
 
 | | Cryo-EM structure of Qb-Ab40 | | Descriptor: | Heavy chain of Ab40, Light chain of Ab40, Minor capsid protein A1 | | Authors: | Bao, K.Y, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
8W5F
 
 | | Cryo-EM structure of Qb-Ab6 | | Descriptor: | Heavy chain of Ab6, Light chain of Ab6, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-26 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
8W5Q
 
 | | Cryo-EM structure of Qb-Ab45 | | Descriptor: | Heavy chain of Ab45, Light chain of Ab45, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
6QA1
 
 | | ERK2 mini-fragment binding | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, pyridin-2-amine | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
5OEM
 
 | |
8W5V
 
 | | Cryo-EM structure of QbN10K-Ab40 | | Descriptor: | Heavy chain of Ab40, Light chain of Ab40, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hua, Z.L, Hou, B.D. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
8W5U
 
 | | Cryo-EM structure of QbN10F-Ab40 | | Descriptor: | Heavy chain of Ab40, Light chain of Ab40, Minor capsid protein A1 | | Authors: | Bao, K.Y, Li, R.H, Hou, B.D, Zhu, P. | | Deposit date: | 2023-08-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Competition propels, rather than limits, the success of low-affinity B cells in the germinal center response.
Cell Rep, 44, 2025
|
|
3C2E
 
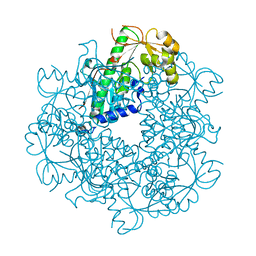 | |
4WUD
 
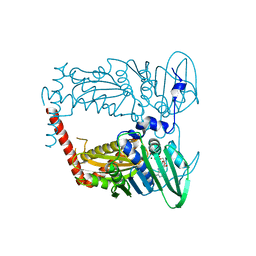 | | N-terminal 43 kDa fragment of the E. coli DNA gyrase B subunit grown from no salt condition | | Descriptor: | CHLORIDE ION, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Hearnshaw, S.J, Chung, T.T, Stevenson, C.E.M, Maxwell, A, Lawson, D.M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The role of monovalent cations in the ATPase reaction of DNA gyrase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8EYZ
 
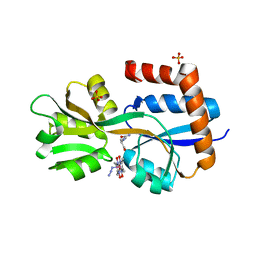 | | Engineered glutamine binding protein bound to GLN and a cobaloxime ligand | | Descriptor: | AZIDOBIS (DIMETHYLGLYOXIMATO) PYRIDINECOBALT, Amino acid ABC transporter substrate-binding protein, GLUTAMINE, ... | | Authors: | Bridwell-Rabb, J, Boggs, D.G, Olshansky, L, Fatima, S, Thompson, P. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Engineering a Conformationally Switchable Artificial Metalloprotein.
J.Am.Chem.Soc., 144, 2022
|
|
7YFK
 
 | | The structure of human pregnane X receptor in complex with an SRC-1 coactivator peptide and a limonoid compound, nomilin | | Descriptor: | Nomilin, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 | | Authors: | Xia, Y, Yao, D, Huang, C, Cao, Y. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pregnane X receptor agonist nomilin extends lifespan and healthspan in preclinical models through detoxification functions.
Nat Commun, 14, 2023
|
|
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
6TDA
 
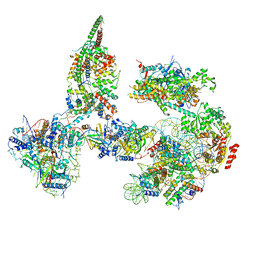 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
6PVA
 
 | | The structure of NTMT1 in complex with compound 11 | | Descriptor: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of NTMT1 in complex with compound 11
To Be published
|
|
6PVJ
 
 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
