8WDF
 
 | | Chemoreceptor PilJ from Pseudomonas aeruginosa PA14 | | Descriptor: | Protein PilJ | | Authors: | Cui, R, Li, D.F. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | The ligand binding domain of a type IV pilus chemoreceptor PilJ has a different fold from that of another PilJ-type receptor McpN.
Biochem.Biophys.Res.Commun., 706, 2024
|
|
8QZH
 
 | | Crystal structure of apo-PptT from Mycobacterium tuberculosis | | Descriptor: | 4'-phosphopantetheinyl transferase PptT, ACETATE ION, MAGNESIUM ION | | Authors: | Gavalda, S, Carivenc, C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Cycle of Type II 4'-Phosphopantetheinyl Transferases
Acs Catalysis, 14, 2024
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7MQE
 
 | | Bartonella henselae NrnC complexed with pAGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
3PSH
 
 | | Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein MolA (MolA bound to Molybdate) | | Descriptor: | MOLYBDATE ION, protein HI_1472 | | Authors: | Tirado-Lee, L, Lee, A, Rees, D.C, Pinkett, H.W. | | Deposit date: | 2010-12-01 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Classification of a Haemophilus influenzae ABC Transporter HI1470/71 through Its Cognate Molybdate Periplasmic Binding Protein, MolA.
Structure, 19, 2011
|
|
7MQB
 
 | | Bartonella henselae NrnC bound to pGG. D4 Symmetry | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
4WMG
 
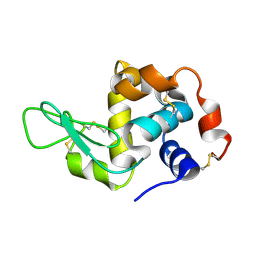 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8QRW
 
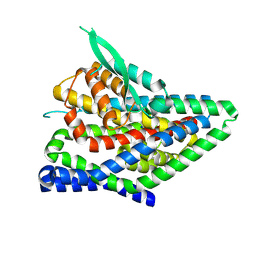 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the intermediate outward-facing state (iOFS-up) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
3PX8
 
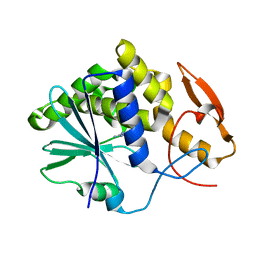 | | RTA in complex with 7-carboxy-pterin | | Descriptor: | 2-amino-4-oxo-1,4-dihydropteridine-7-carboxylic acid, Preproricin | | Authors: | Jasheway, K.R, Robertus, J.D. | | Deposit date: | 2010-12-09 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 7-Substituted pterins provide a new direction for ricin A chain inhibitors.
Eur.J.Med.Chem., 46, 2011
|
|
6T41
 
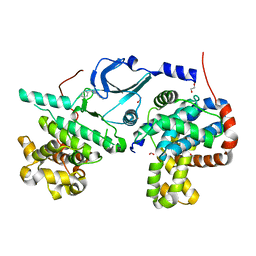 | | CDK8/Cyclin C in complex with N-(4-chlorobenzyl)isoquinolin-4-amine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Maskos, K, Huber, R, Kuhn, C.-D. | | Deposit date: | 2019-10-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A precisely positioned MED12 activation helix stimulates CDK8 kinase activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RZW
 
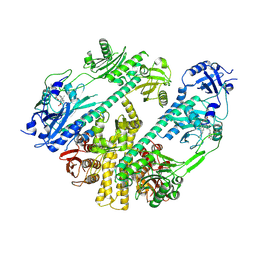 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
8QRS
 
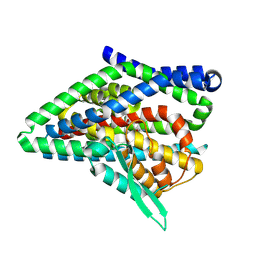 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-up) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
8QRU
 
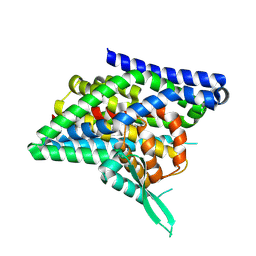 | | ASCT2 protomer in lipid nanodiscs with bound glutamine and Na+ ions in the intermediate outward-facing state (iOFS-down) | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
7SD0
 
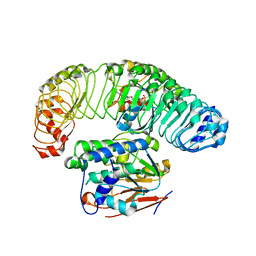 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
8BSC
 
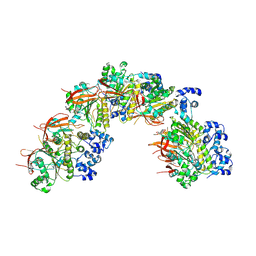 | |
7MUQ
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 1 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
8W35
 
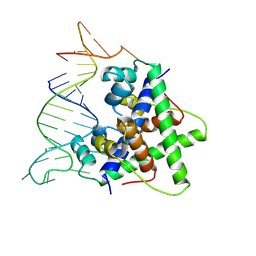 | | Aca2 from Pectobacterium phage ZF40 bound to RNA | | Descriptor: | Anti-CRISPR associated (Aca) protein, Aca2, IR2 and IR-RBS RNA | | Authors: | Wilkinson, M.E, Birkholz, N, Kimanius, D, Fineran, P.C. | | Deposit date: | 2024-02-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Phage anti-CRISPR control by an RNA- and DNA-binding helix-turn-helix protein.
Nature, 631, 2024
|
|
6TEA
 
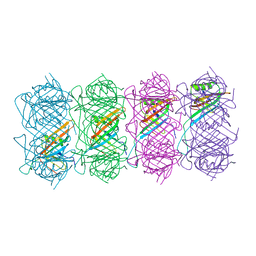 | | Tail of native GTA particle computed with helical refinement, C6 symmetry | | Descriptor: | Phage major tail protein, TP901-1 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
8QRV
 
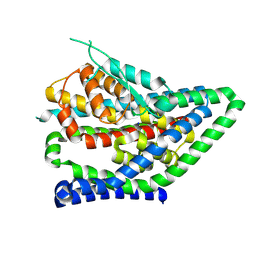 | | ASCT2 protomer in lipid nanodiscs under low Na+ concentration in the outward-facing state (OFS) | | Descriptor: | Neutral amino acid transporter B(0), SODIUM ION | | Authors: | Borowska, A, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of the obligatory exchange mode of human neutral amino acid transporter ASCT2.
Nat Commun, 15, 2024
|
|
5E03
 
 | | Crystal structure of mouse CTLA-4 nanobody | | Descriptor: | CTLA-4 nanobody, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Crystal structure of mouse CTLA-4 nanobody
To Be Published
|
|
7MUW
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 4 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
5E0Z
 
 | |
8HFK
 
 | |
7MUS
 
 | | Reconstruction of the Legionella pneumophila Dot/Icm T4SS 3DVA Map 2 | | Descriptor: | DUF2807 domain-containing protein, DotC, DotD, ... | | Authors: | Sheedlo, M.J, Durie, C.L, Swanson, M, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2021-05-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS.
Elife, 10, 2021
|
|
8HFJ
 
 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | Descriptor: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Yin, D.J, Rao, Y.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
