8PAB
 
 | |
8PIZ
 
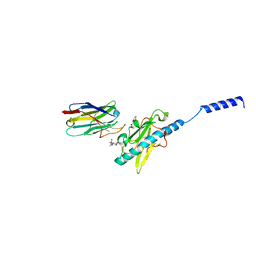 | | Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody | | Descriptor: | 2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, C24 nanobody, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
7P84
 
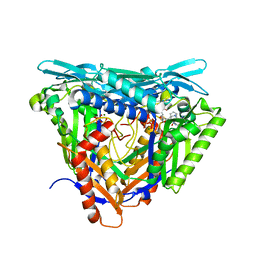 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P8M
 
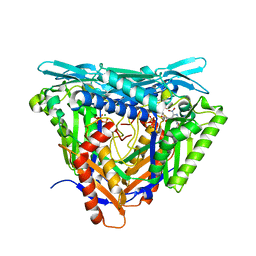 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with DMNB-SAM (4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | 4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine, MAGNESIUM ION, S-adenosylmethionine synthase, ... | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P82
 
 | | Crystal structure of apo form L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
6O6R
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, AMTB-bound state | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, CHOLESTEROL HEMISUCCINATE, N-(3-aminopropyl)-2-[(3-methylphenyl)methoxy]-N-[(thiophen-2-yl)methyl]benzamide, ... | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
6UK1
 
 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
7PBW
 
 | | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Qian, P, Swainsbury, D.J.K, Croll, T.I, Castro-Hartmann, P, Sader, K, Divitini, G, Hunter, C.N. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Cryo-EM Structure of the Rhodobacter sphaeroides Light-Harvesting 2 Complex at 2.1 angstrom.
Biochemistry, 60, 2021
|
|
7P83
 
 | | Crystal structure of Apo form of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
4X7I
 
 | | Crystal Structure of BACE with amino thiazine inhibitor LY2886721 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(4aS,7aS)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Timm, D.E. | | Deposit date: | 2014-12-09 | | Release date: | 2014-12-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Potent BACE1 Inhibitor LY2886721 Elicits Robust Central A beta Pharmacodynamic Responses in Mice, Dogs, and Humans.
J.Neurosci., 35, 2015
|
|
8VVB
 
 | | Influenza antibody L5A7 Fab | | Descriptor: | L5A7 Heavy Chain, L5A7 Light Chain | | Authors: | Harris, D.R, Olia, A.S, Kwong, P.D. | | Deposit date: | 2024-01-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody.
Front Immunol, 15, 2024
|
|
6WBW
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5MCB
 
 | | Glycogen phosphorylase in complex with chlorogenic acid. | | Descriptor: | Chlorogenic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Leonidas, D.D, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Kantsadi, A.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Affinity Crystallography Reveals the Bioactive Compounds of Industrial Juicing Byproducts of Punica granatum for Glycogen Phosphorylase.
Curr Drug Discov Technol, 15, 2018
|
|
6TK3
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 30us+150us structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TKH
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - orthorhombic form at 7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SODIUM ION, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
6TD3
 
 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | Descriptor: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
5MQS
 
 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
6WKD
 
 | | Crystal structure of pentalenene synthase complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
7S97
 
 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
5L8Y
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-937 | | Descriptor: | 2-[(4-{5-[(4aR,8aS)-3-cycloheptyl-4-oxo-3,4,4a,5,8,8a-hexahydrophthalazin-1-yl]-2-methoxyphenyl}phenyl)formamido]-N-(2-hydroxyethyl)acetamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-08 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
6YX2
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[(~{E})-5-oxidanylidenepentanoyldiazenyl]methyl]benzoic acid, PWW-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
7LY0
 
 | |
6ARU
 
 | | Structure of Cetuximab Fab mutant in complex with EGFR extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab mutant heavy chain Fab fragment,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Christie, M, Christ, D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Cetuximab Fab mutant in complex with EGFR extracellular domain
To Be Published
|
|
