1D7R
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 2,2-DIALKYLGLYCINE DECARBOXYLASE WITH 5PA | | Descriptor: | N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
4CWT
 
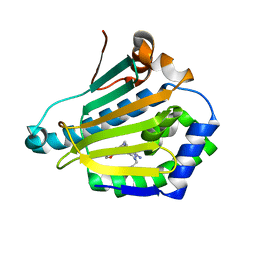 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-{[(2Z)-5-(1,3-benzodioxol-5-ylmethyl)-8-fluoro-2-imino-2,3-dihydro[1,2,4]triazolo[1,5-c]quinazolin-10-yl]amino}ethanol, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4CWQ
 
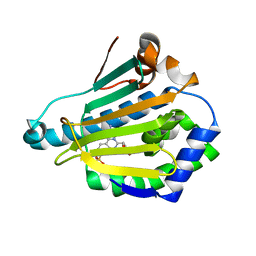 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 2-amino-5-(1,3-benzodioxol-5-ylmethyl)[1,2,4]triazolo[1,5-c]quinazoline-8-sulfonamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
5JHU
 
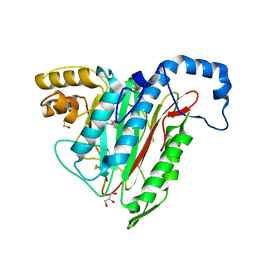 | | Potent, Reversible MetAP2 Inhibitors via FBDD | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 1.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7NWK
 
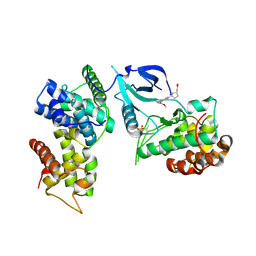 | | Crystal structure of CDK9-Cyclin T1 bound by compound 6 | | Descriptor: | Cyclin-T1, Cyclin-dependent kinase 9, N-((1R,3R)-3-(7-(4-fluoro-2-methoxyphenyl)-3H-imidazo[4,5-b]pyridin-2-yl)cyclopentyl)acetamide | | Authors: | Collie, G.W, Ferguson, A.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of a Series of 7-Azaindoles as Potent and Highly Selective CDK9 Inhibitors for Transient Target Engagement.
J.Med.Chem., 64, 2021
|
|
6ENF
 
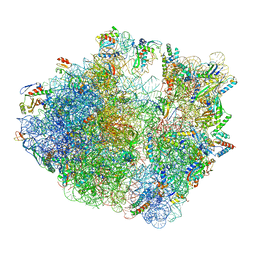 | | Cryo-EM structure of a polyproline-stalled ribosome in the absence of EF-P | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-04 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
5J3R
 
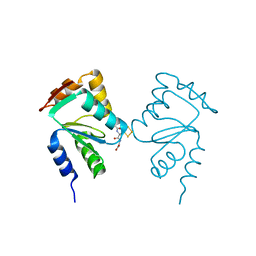 | | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Abdalla, M, Dai, Y.-N, Chi, C.-B, Cheng, W, Cao, D.-D, Zhou, K, Ali, W, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2016-03-31 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JMF
 
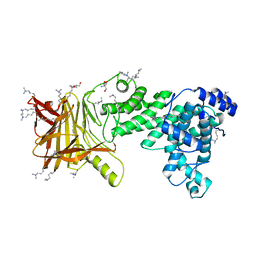 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
1TOT
 
 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1D31
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
5J31
 
 | | Crystal structure of 14-3-3zeta in complex with an alkyne cross-linked cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Exoenzyme S | | Authors: | Wallraven, K, Cromm, P, Bier, D, Glas, A, Grossmann, T. | | Deposit date: | 2016-03-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis.
Chembiochem, 17, 2016
|
|
1D7U
 
 | | Crystal structure of the complex of 2,2-dialkylglycine decarboxylase with LCS | | Descriptor: | POTASSIUM ION, PROTEIN (2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE)), SODIUM ION, ... | | Authors: | Malashkevich, V.N, Toney, M.D, Strop, P, Keller, J, Jansonius, J.N. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of dialkylglycine decarboxylase inhibitor complexes.
J.Mol.Biol., 294, 1999
|
|
1D92
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH G.T MISMATCHED BASE-PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*TP*CP*C)-3') | | Authors: | Hunter, W.N, Kneale, G, Brown, T, Rabinovich, D, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with G . T mismatched base-pairs.
J.Mol.Biol., 190, 1986
|
|
7OB9
 
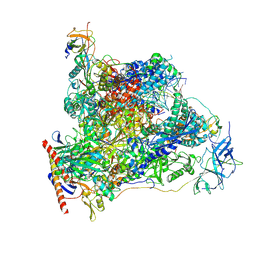 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBA
 
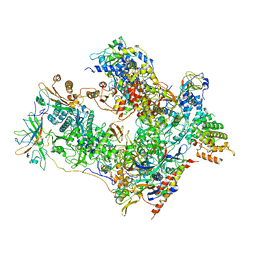 | | Cryo-EM structure of human RNA Polymerase I in complex with RRN3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6DZA
 
 | |
6E5H
 
 | |
1TTK
 
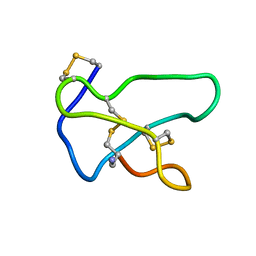 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
4US8
 
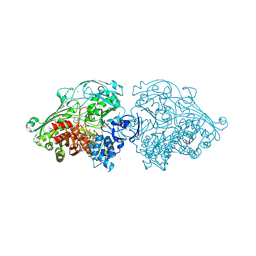 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with benzaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
1TTG
 
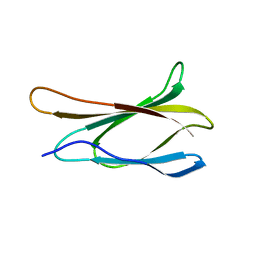 | | THE THREE-DIMENSIONAL STRUCTURE OF THE TENTH TYPE III MODULE OF FIBRONECTIN: AN INSIGHT INTO RGD-MEDIATED INTERACTIONS | | Descriptor: | FIBRONECTIN | | Authors: | Main, A.L, Harvey, T.S, Baron, M, Campbell, I.D. | | Deposit date: | 1993-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Cell(Cambridge,Mass.), 71, 1992
|
|
1TVI
 
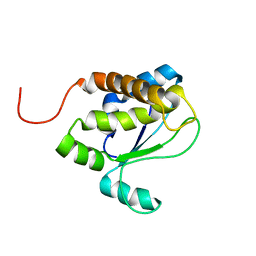 | | Solution structure of TM1509 from Thermotoga maritima: VT1, a NESGC target protein | | Descriptor: | Hypothetical UPF0054 protein TM1509 | | Authors: | Penhoat, C.H, Atreya, H.S, Kim, S, Li, Z, Yee, A, Xiao, R, Murray, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure.
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1TR6
 
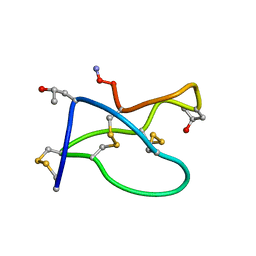 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2011-10-05 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
4DK2
 
 | |
1U27
 
 | | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,3,4,5)P4 | | Descriptor: | Cytohesin 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
