2IHD
 
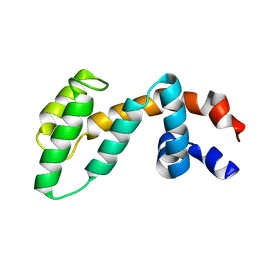 | | Crystal structure of Human Regulator of G-protein signaling 8, RGS8 | | Descriptor: | CHLORIDE ION, Regulator of G-protein signaling 8 | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Salah, E, Gileadi, C, Burgess, N, Bhatia, C, Gileadi, O, Bray, J, Elkins, J, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6W2I
 
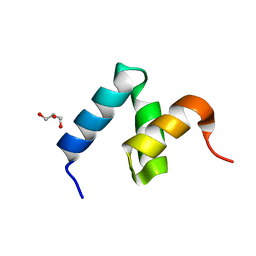 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A | | Descriptor: | GLYCEROL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
3ZTF
 
 | | X-ray Structure of the Cyan Fluorescent Protein mTurquoise2 (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
4R0W
 
 | |
4RHI
 
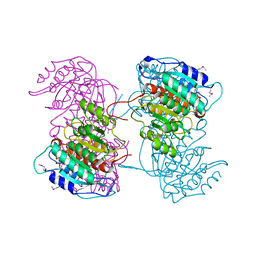 | |
4RHB
 
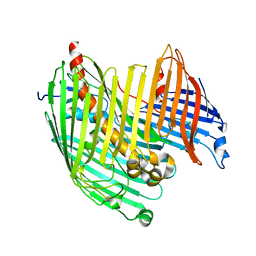 | |
4RHL
 
 | |
4RHQ
 
 | | Crystal structure of T. brucei arginase-like protein double mutant S149D/S153D | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL | | Authors: | Hai, Y, Barrett, M.P, Christianson, D.W. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
4RHM
 
 | | Crystal structure of T. brucei arginase-like protein quadruple mutant S149D/R151H/S153D/S226D | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Barrett, M.P, Christianson, D.W. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Arginase-like Protein from Trypanosoma brucei That Evolved without a Binuclear Manganese Cluster.
Biochemistry, 54, 2015
|
|
4R3S
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb6D8)in complex with MSP2 11-23 | | Descriptor: | FV FRAGMENT(MAB6D8) HEAVY CHAIN, FV FRAGMENT(MAB6D8) LIGHT CHAIN, Merozoite surface protein | | Authors: | Morales, R.A.V, MacRaild, C.A, Seow, J, Bankala, K, Drinkwater, N, McGowan, S, Rouet, R, Christ, D, Anders, R.F, Norton, R.S. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for epitope masking and strain specificity of a conserved epitope in an intrinsically disordered malaria vaccine candidate
Sci Rep, 5, 2015
|
|
4R5I
 
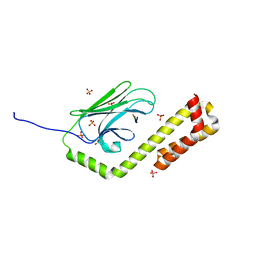 | | Crystal structure of the DnaK C-terminus with the substrate peptide NRLLLTG | | Descriptor: | Chaperone protein DnaK, HSP70/DnaK Substrate Peptide: NRLLLTG, PHOSPHATE ION, ... | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9702 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
2ILU
 
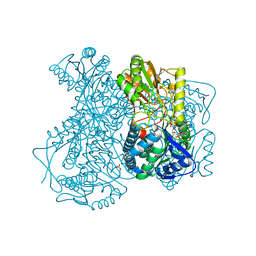 | |
2IED
 
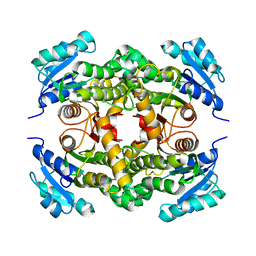 | | CRYSTAL STRUCTURE of ISONIAZID-RESISTANT S94A ENOYL-ACP(COA) REDUCTASE MUTANT ENZYME FROM MYCOBACTERIUM TUBERCULOSIS UNCOMPLEXED | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Dias, M.V.B, Prado, A.M.X, Vasconcelos, I.B, Fadel, V, Basso, L.A, Santos, D.S, Azevedo Jr, W.F. | | Deposit date: | 2006-09-18 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic studies on the binding of isonicotinyl-NAD adduct to wild-type and isoniazid resistant 2-trans-enoyl-ACP (CoA) reductase from Mycobacterium tuberculosis.
J.Struct.Biol., 159, 2007
|
|
4RJL
 
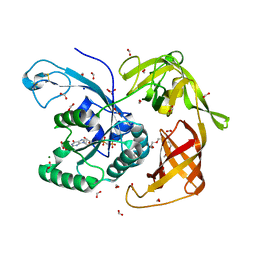 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kravchenko, O.V, Nikonov, O.S, Arhipova, V.I, Stolboushkina, E.A, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2014-10-09 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Crystal structure of gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDPCP at 1.64A resolution
To be Published
|
|
6WBU
 
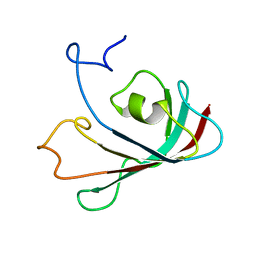 | |
2II8
 
 | | Anabaena sensory rhodopsin transducer | | Descriptor: | Anabaena sensory rhodopsin transducer protein | | Authors: | Vogeley, L, Sineshchekov, O.A, Trivedi, V.D, Spudich, E.N, Spudich, J.L, Luecke, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anabaena sensory rhodopsin transducer.
J.Mol.Biol., 367, 2007
|
|
7T1L
 
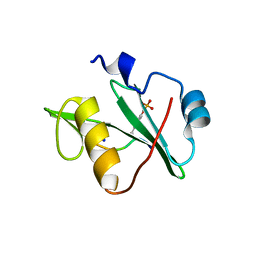 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
4RSP
 
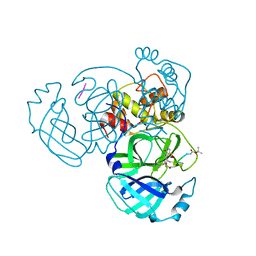 | |
3EVS
 
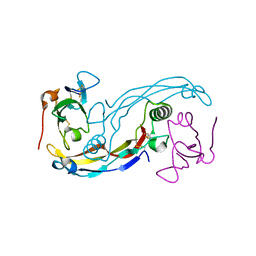 | | Crystal structure of the GDF-5:BMP receptor IB complex. | | Descriptor: | Bone morphogenetic protein receptor type-1B, Growth/differentiation factor 5 | | Authors: | Kotzsch, A, Mueller, T.D. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity.
Embo J., 28, 2009
|
|
3EZE
 
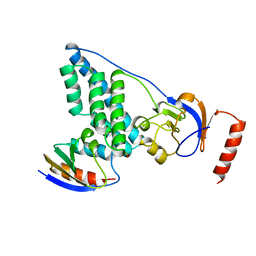 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHITE ION, PROTEIN (PHOSPHOTRANSFERASE SYSTEM, ENZYME I), ... | | Authors: | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
4S1Z
 
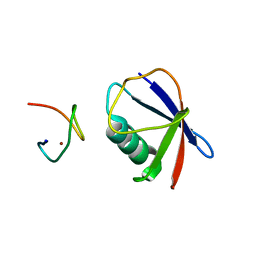 | | Crystal structure of TRABID NZF1 in complex with K29 linked di-Ubiquitin | | Descriptor: | Ubiquitin, Ubiquitin thioesterase ZRANB1, ZINC ION | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Campbell, D.G, Morrice, N.A, Johnson, C, Toth, R, Kulathu, Y. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin.
Mol.Cell, 58, 2015
|
|
6J2D
 
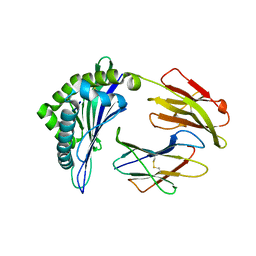 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | HeV1, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6JIU
 
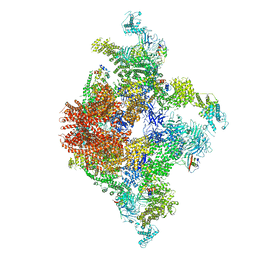 | | Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
5K6C
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 variant. | | Descriptor: | Fusion glycoprotein F0,Fusion glycoprotein F0, SULFATE ION | | Authors: | Joyce, M.G, Zhang, B, Lai, Y.T, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-12 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (3.576 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
