6I7M
 
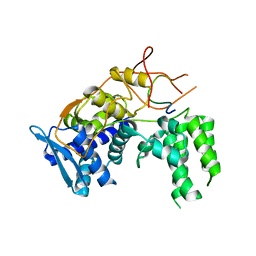 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6YKI
 
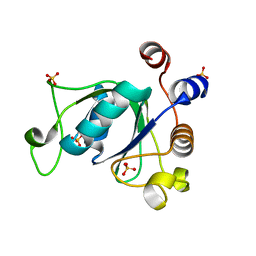 | | Crystal structure of YTHDC1 with compound DHU_DC1_092 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-ethyl-2-[(2~{S},5~{R})-5-methyl-2-phenyl-morpholin-4-yl]ethanamine | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
8AWW
 
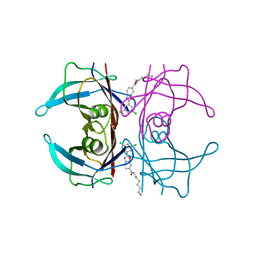 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3JYP
 
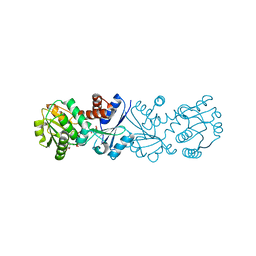 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
6P8D
 
 | |
6I69
 
 | |
6YKE
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_038 | | Descriptor: | (2~{R})-2-(3-fluorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6PBE
 
 | | ZINC17988990-bound TRPV5 in nanodiscs | | Descriptor: | (4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)methyl 3-(3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)propanoate, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
6YL0
 
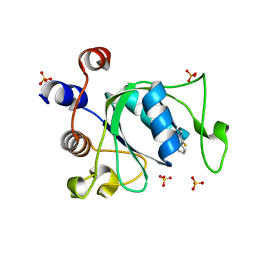 | | Crystal structure of YTHDC1 with compound T_96 | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of YTHDC1 with compound T_96
To Be Published
|
|
6YL9
 
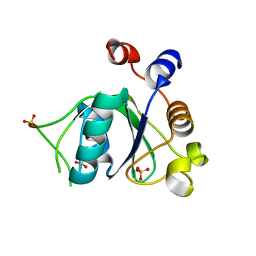 | | Crystal structure of YTHDC1 with compound DHU_DC1_085 | | Descriptor: | 3-[(2~{R},5~{S})-2-(2,5-dimethylphenyl)-5-methyl-morpholin-4-yl]propane-1-sulfonamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7R1O
 
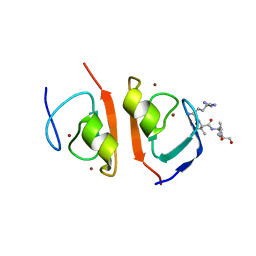 | | p62-ZZ domain of the human sequestosome in complex with dusquetide | | Descriptor: | Dusquetide, Sequestosome-1, ZINC ION | | Authors: | Hakansson, M, Hansson, M, Logan, D.T, Rozek, A, Donini, O. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Dusquetide modulates innate immune response through binding to p62.
Structure, 30, 2022
|
|
8G95
 
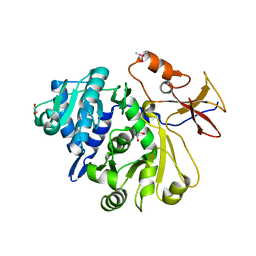 | |
5J6Q
 
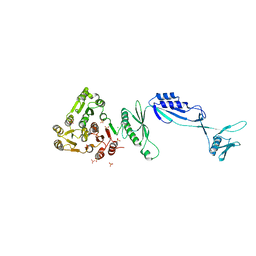 | | Cwp8 from Clostridium difficile | | Descriptor: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
8AOL
 
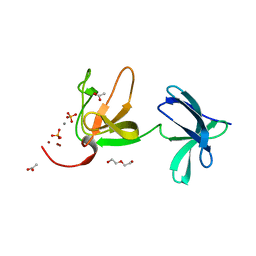 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain III (aa 363-499) | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sagmeister, T, Damisch, E, Eder, M, Dordic, A, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6Y2X
 
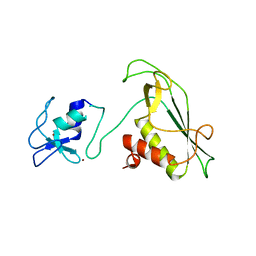 | | RING-DTC domains of Deltex 2, Form 2 | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
3JBP
 
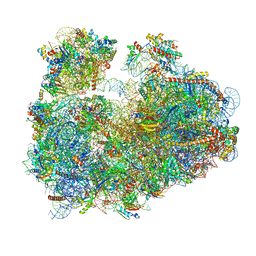 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
6Y2Z
 
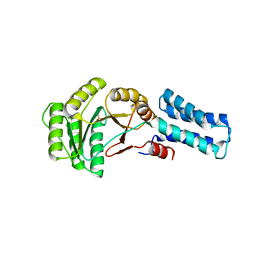 | | NG domain of human SRP54 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Signal recognition particle 54 kDa protein | | Authors: | Juaire, K.D, Wild, K, Sinning, I. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia.
Structure, 29, 2021
|
|
6V3I
 
 | |
8G5O
 
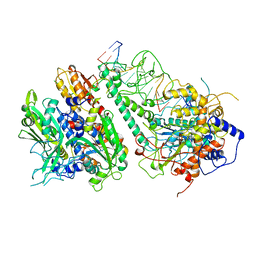 | | Cryo-EM structure of the Guide loop Engagement Complex (IV) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
6LNT
 
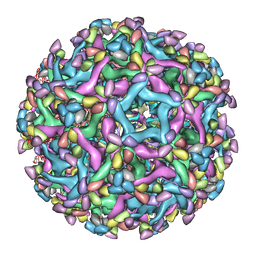 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
5JB1
 
 | | Pseudo-atomic structure of Human Papillomavirus Type 59 L1 Virus-like Particle | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Zheng, Q.B, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
5JBN
 
 | |
7RAD
 
 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-1-(4-phenylphenyl)-6,7,8,9-tetrahydro-5~{H}-imidazo[1,2-a][1,3]diazepine, Aldehyde dehydrogenase X, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-07-01 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
8FWO
 
 | | Crystal structure of SARS-CoV-2 papain-like protease | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease
To Be Published
|
|
6YA7
 
 | | Cdc7-Dbf4 bound to an Mcm2-S40 derived bivalent substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, DNA replication licensing factor MCM2, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
