8FCP
 
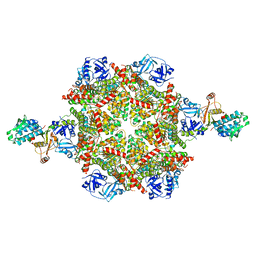 | | Cryo-EM structure of p97:UBXD1 para state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8UCI
 
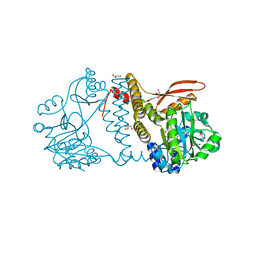 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
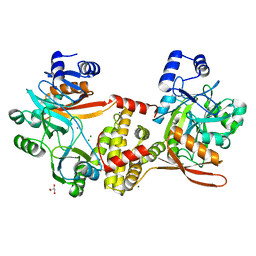 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
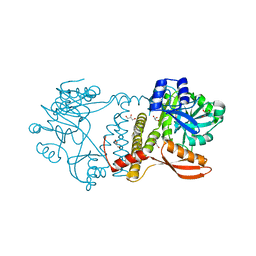 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
2MBK
 
 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | Descriptor: | Crossveinless 2 | | Authors: | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | Deposit date: | 2013-08-02 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
8FCQ
 
 | | Cryo-EM structure of p97:UBXD1 PUB-in state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2MA5
 
 | | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7375C | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Hassan, F, Ramelot, T.A, Yang, Y, Cort, J.R, Janjua, H, Kohan, E, Lee, D, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific
demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural
Genomics Consortium (NESG) Target HR7375C
To be Published
|
|
6VU5
 
 | | Structure of G-alpha-q bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(q) subunit alpha, Resistance to inhibitors of cholinesterase-8A (Ric-8A) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
8FCM
 
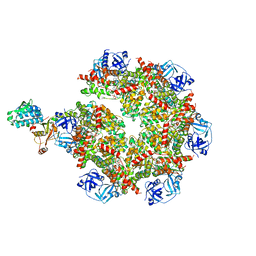 | | Cryo-EM structure of p97:UBXD1 open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2MBL
 
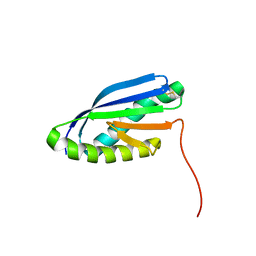 | | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33 | | Descriptor: | Top7 Fold Protein Top7m13 | | Authors: | Liu, G, Zanghellini, A.L, Chan, K, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-08-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Top7 Fold Protein Top7m13, Northeast Structural Genomics Consortium (NESG) Target OR33
To be Published
|
|
2G2B
 
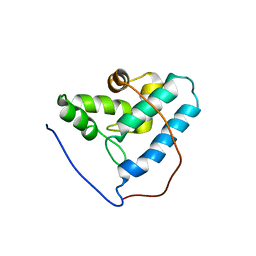 | | NMR structure of the human allograft inflammatory factor 1 | | Descriptor: | Allograft inflammatory factor 1 | | Authors: | Song, J, Tyler, R.C, Newman, C.L, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-15 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human allograft inflammatory factor 1
To be published
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
2M7P
 
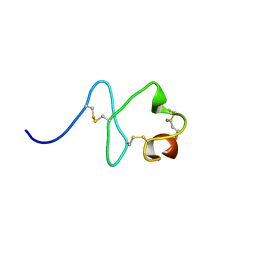 | | RXFP1 utilises hydrophobic moieties on a signalling surface of the LDLa module to mediate receptor activation | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Relaxin receptor 1 | | Authors: | Kong, R.CK, Petrie, E.J, Mohanty, B, Ling, J, Lee, J.C.Y, Gooley, P.R, Bathgate, R.A.D. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.
J.Biol.Chem., 288, 2013
|
|
5J72
 
 | | Cwp6 from Clostridium difficile | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
2MJ5
 
 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | Descriptor: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-12-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
6DLU
 
 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
2LYJ
 
 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
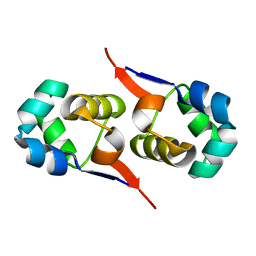 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2M1P
 
 | | [Aba5,14]BTD-2 | | Descriptor: | [Aba5,14]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
3GOP
 
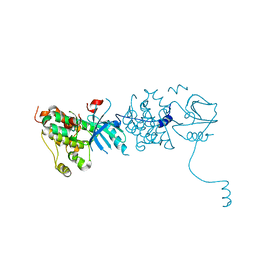 | |
2M9Y
 
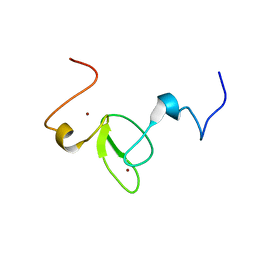 | |
2M9X
 
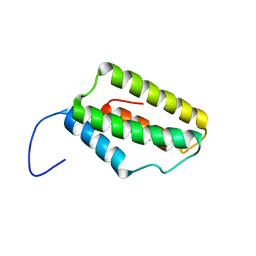 | | Solution NMR Structure of Microtubule-associated serine/threonine-protein kinase 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9151A | | Descriptor: | Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Xu, X, Eletsky, A, Shastry, R, Lee, D, Hamilton, K, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Microtubule-associated serine/threonine-protein kinase 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9151A
To be Published
|
|
1AT9
 
 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
3FM1
 
 | | Crystal Structure Analysis of Fungal Versatile Peroxidase from Pleurotus eryngii | | Descriptor: | CALCIUM ION, FE (III) ION, MANGANESE (II) ION, ... | | Authors: | Piontek, K, Martinez, A.T, Choinowski, T, Plattner, D.A. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and Site-directed Mutagenesis Study of Versatile Peroxidase Oxidizing both Mn(II) and Aromatic Substrates
To be Published
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
