3F3T
 
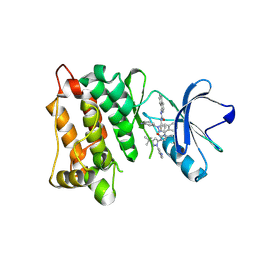 | | Kinase domain of cSrc in complex with inhibitor RL38 (Type III) | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Klueter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-03-03 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new screening assay for allosteric inhibitors of cSrc
Nat.Chem.Biol., 5, 2009
|
|
6ZKB
 
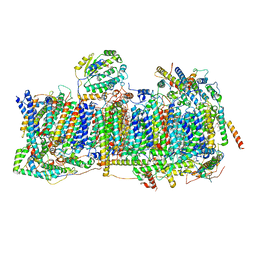 | | Membrane domain of closed complex I during turnover | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
9BX6
 
 | | Class 9 model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
2J53
 
 | | Solution Structure of GB1 domain Protein G and low and high pressure. | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wilton, D.J, Tunnicliffe, R.B, Kamatari, Y.O, Akasaka, K, Williamson, M.P. | | Deposit date: | 2006-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pressure-Induced Changes in the Solution Structure of the Gb1 Domain of Protein G.
Proteins, 71, 2008
|
|
9BX9
 
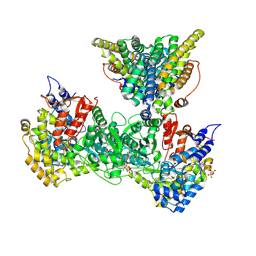 | | Class 15 model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
9BX8
 
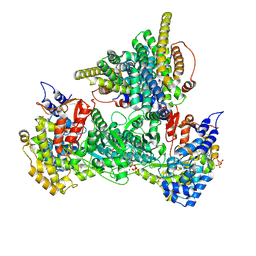 | | Class 12 model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
3FAY
 
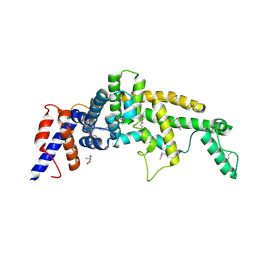 | | Crystal structure of the GAP-related domain of IQGAP1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ras GTPase-activating-like protein IQGAP1 | | Authors: | Kurella, V.B, Richard, J.M, Parke, C.L, Bellamy, H, Worthylake, D.K. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the GTPase-activating protein-related domain from IQGAP1.
J.Biol.Chem., 284, 2009
|
|
9BWX
 
 | | Consensus full-complex model for preturnover condition of Bacillus subtilis ribonucleotide reductase complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Xu, D, Thomas, W.C, Burnim, A.A, Ando, N. | | Deposit date: | 2024-05-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Conformational landscapes of a class I ribonucleotide reductase complex during turnover reveal intrinsic dynamics and asymmetry.
Nat Commun, 16, 2025
|
|
3FB5
 
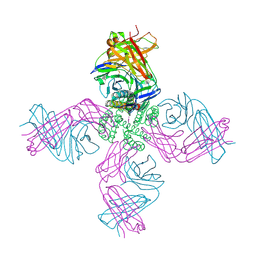 | | KcsA potassium channel in the partially open state with 14.5 A opening at T112 | | Descriptor: | POTASSIUM ION, Voltage-gated potassium channel, antibody Fab fragment heavy chain, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Perozo, E. | | Deposit date: | 2008-11-18 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KcsA potassium channel in the partially open state with 14.5 A opening at T112
TO BE PUBLISHED
|
|
3FBM
 
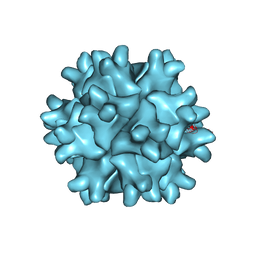 | | D431N Mutant VP2 Protein of Infectious Bursal Disease Virus; Derived T=1 Particles | | Descriptor: | CALCIUM ION, Polyprotein | | Authors: | Irigoyen, N, Garriga, D, Navarro, A, Verdaguer, N, Rodriguez, J.F, Caston, J.R. | | Deposit date: | 2008-11-19 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoproteolytic Activity Derived from the Infectious Bursal Disease Virus Capsid Protein
J.Biol.Chem., 284, 2009
|
|
2FS1
 
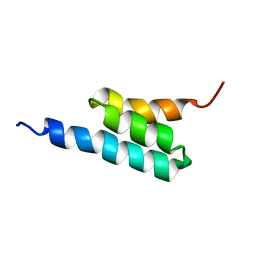 | | solution structure of PSD-1 | | Descriptor: | PSD-1 | | Authors: | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2006-01-20 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
9BS1
 
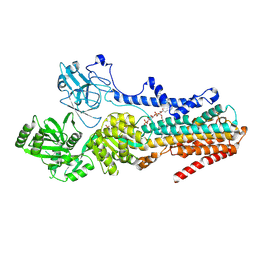 | | Cryo-EM structure of the S. cerevisiae lipid flippase Neo1 bound with PI4P in the E2P state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl di[(9Z)-octadec-9-enoate], BERYLLIUM TRIFLUORIDE ION, Phospholipid-transporting ATPase NEO1 | | Authors: | Duan, H.D, Li, H. | | Deposit date: | 2024-05-12 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | P4-ATPase control over phosphoinositide membrane asymmetry and neomycin resistance.
Biorxiv, 2025
|
|
2J4M
 
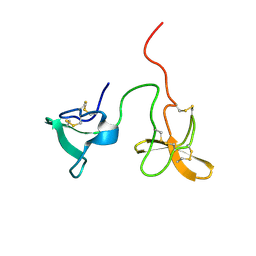 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
8CRS
 
 | |
9BBV
 
 | |
7AIL
 
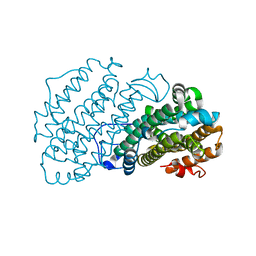 | |
9CYP
 
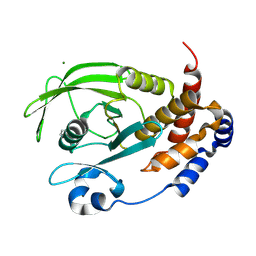 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2025
|
|
6VU8
 
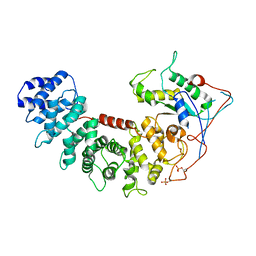 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
2J2S
 
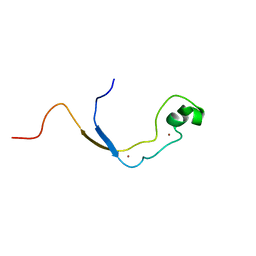 | | Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase | | Descriptor: | ZINC FINGER PROTEIN HRX, ZINC ION | | Authors: | Allen, M.D, Grummitt, C.G, Hilcenko, C, Young-Min, S, Tonkin, L.M, Johnson, C.M, Bycroft, M, Warren, A.J. | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nonmethyl-Cpg-Binding Cxxc Domain of the Leukaemia-Associated Mll Histone Methyltransferase
Embo J., 25, 2006
|
|
3F6Q
 
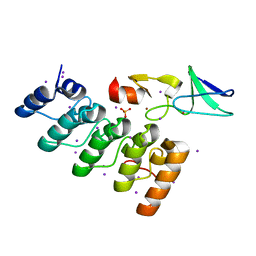 | | Crystal structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain | | Descriptor: | IODIDE ION, Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Chiswell, B.P, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of integrin-linked kinase-PINCH interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8F7B
 
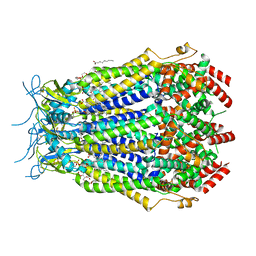 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9CYO
 
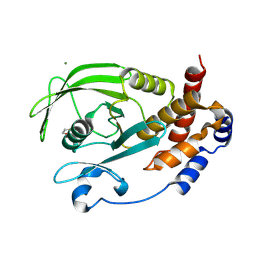 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2025
|
|
8F79
 
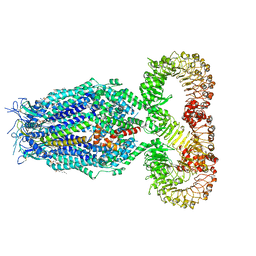 | | LRRC8A(T48D):C conformation 2 top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9CYQ
 
 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2025
|
|
8QNO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase treated at 368 K from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Koeppl, L.H, Popadic, D, Andexer, J.N. | | Deposit date: | 2023-09-27 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
