6E6U
 
 | | Variant C89S of Dieckmann cyclase, NcmC | | Descriptor: | Dieckmann cyclase, NcmC, SULFATE ION | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Enzymatic Off-Loading of Hybrid Polyketides by Dieckmann Condensation.
Acs Chem.Biol., 2020
|
|
6E8F
 
 | |
6EB5
 
 | |
4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | Authors: | Williams, L.K, Caner, S, Brayer, G.D. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
1D8Y
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA | | Descriptor: | D(T)19 OLIGOMER, DNA POLYMERASE I, SULFATE ION, ... | | Authors: | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | Deposit date: | 1999-10-26 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CI0
 
 | | PNP OXIDASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (PNP OXIDASE) | | Authors: | Shi, W, Ostrov, D.A, Gerchman, S.E, Graziano, V, Kycia, H, Studier, B, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-04-06 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of PNP Oxidase from S. Cerevisiae
To be Published
|
|
6EI2
 
 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
2OLW
 
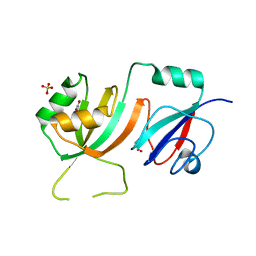 | | Crystal Structure of E. coli pseudouridine synthase RluE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETIC ACID, Ribosomal large subunit pseudouridine synthase E, ... | | Authors: | Pan, H, Ho, J.D, Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of E. coli rRNA Pseudouridine Synthase RluE.
J.Mol.Biol., 367, 2007
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
4CVA
 
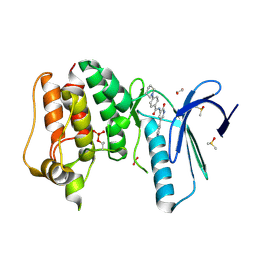 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4V11
 
 | | Structure of Synaptotagmin-1 with SV2A peptide phosphorylated at Thr84 | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTIC VESICLE GLYCOPROTEIN 2A, ... | | Authors: | Zhang, N, Gordon, S.L, Fritsch, M.J, Esoof, N, Campbell, D, Gourlay, R, Velupillai, S, Macartney, T, Peggie, M, vanAalten, D.M.F, Cousin, M.A, Alessi, D.R. | | Deposit date: | 2014-09-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
J.Neurosci., 35, 2015
|
|
4CO8
 
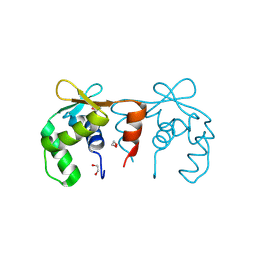 | | Structure of the DNA binding ETS domain of human ETV4 | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 4 | | Authors: | Newman, J.A, Cooper, C.D.O, Shrestha, L, Burgess-Brown, N, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
6EH5
 
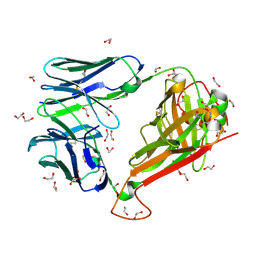 | |
1CNZ
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | MANGANESE (II) ION, PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE), SULFATE ION | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-24 | | Release date: | 1999-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of Eschericia Coli and Salmonella Typhimurium 3-
Isopropylmalate Dehydrogenase and Comparison with Their Thermophilic
Counterpart from Thermus Thermophilus
J.Mol.Biol., 266, 1997
|
|
6Z9C
 
 | | Structure of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability | | Descriptor: | Polymerase delta-interacting protein 2, SODIUM ION | | Authors: | Kulik, A.A, Maruszczak, K, Nabi, N.L.M, Bingham, R.J, Cooper, C.D.O. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and molecular dynamics of human POLDIP2, a multifaceted adaptor protein in metabolism and genome stability.
Protein Sci., 30, 2021
|
|
4LPS
 
 | | Crystal structure of HypB from Helicobacter pylori in complex with nickel | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Hydrogenase/urease nickel incorporation protein HypB, ... | | Authors: | Lebrette, H, Sydor, A.M, Ariyakumaran, R, Zamble, D.B, Cavazza, C. | | Deposit date: | 2013-07-16 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Relationship between Ni(II) and Zn(II) Coordination and Nucleotide Binding by the Helicobacter pylori [NiFe]-Hydrogenase and Urease Maturation Factor HypB.
J.Biol.Chem., 289, 2014
|
|
3AB1
 
 | | Crystal Structure of Ferredoxin NADP+ Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Muraki, N, Seo, D, Kurisu, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+
J.Mol.Biol., 401, 2010
|
|
4D4V
 
 | | Focal Adhesion Kinase catalytic domain | | Descriptor: | 6-methyl-4-(piperazin-1-yl)-2-(trifluoromethyl)quinoline, DIMETHYL SULFOXIDE, FOCAL ADHESION KINASE, ... | | Authors: | Le Coq, J, Lin, A, Lietha, D. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric Regulation of Focal Adhesion Kinase by Pip2 and ATP.
Biophys.J., 108, 2015
|
|
6EOC
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
4CNK
 
 | | L-Aminoacetone oxidase from Streptococcus oligofermentans belongs to a new 3-domain family of bacterial flavoproteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-AMINO ACID OXIDASE, ... | | Authors: | Molla, G, Nardini, M, Motta, P, D'Arrigo, P, Bolognesi, M, Pollegioni, L. | | Deposit date: | 2014-01-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminoacetone Oxidase from Streptococcus Oligofermentas Belongs to a New Three-Domain Family of Bacterial Flavoproteins.
Biochem.J., 464, 2014
|
|
2OPD
 
 | | Structure of the Neisseria meningitidis minor Type IV pilin, PilX | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4COH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the sulfonamide derivative of the 4-aminopyridyl-based inhibitor | | Descriptor: | 2-fluoranyl-N-[(2R)-3-(1H-indol-3-yl)-1-oxidanylidene-1-(pyridin-4-ylamino)propan-2-yl]-4-(4-thiophen-2-ylsulfonylpiperazin-1-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Vieira, D.F, Choi, J.Y, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Expanding the Binding Envelope of Cyp51 Inhibitors Targeting Trypanosoma Cruzi with 4-Aminopyridyl-Based Sulfonamide Derivatives
Chembiochem, 15, 2014
|
|
2OPX
 
 | |
5XI4
 
 | | BRD4 bound with compound Bdi4 | | Descriptor: | (3~{S})-4-cyclopropyl-1,3-dimethyl-6-[[(1~{S})-1-(4-methylphenyl)ethyl]amino]-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2017-04-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | BRD4 bound with compound Bdi4
To Be Published
|
|
6ZB8
 
 | | Exo-beta-1,3-glucanase from moose rumen microbiome, active site mutant E167Q/E295Q | | Descriptor: | Exo-beta-1,3-glucanase variant E167Q/E295Q, POLYETHYLENE GLYCOL (N=34) | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
