2HGK
 
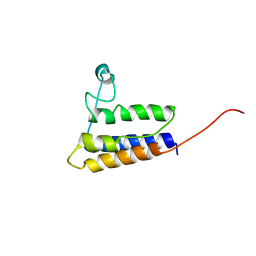 | | Solution NMR Structure of protein YqcC from E. coli: Northeast Structural Genomics Consortium target ER225 | | Descriptor: | Hypothetical protein yqcC | | Authors: | Cort, J.R, Wang, D, Shetty, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-27 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YqcC from E.coli
To be Published
|
|
1ROD
 
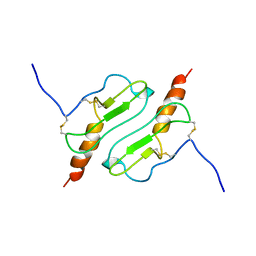 | | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN, NMR | | Descriptor: | CHIMERIC PROTEIN OF INTERLEUKIN 8 AND HUMAN MELANOMA GROWTH STIMULATING ACTIVITY PROTEIN | | Authors: | Roesch, P, Sticht, H, Auer, M, Schmitt, B, Besemer, J, Horcher, M, Kirsch, T, Lindley, I.J.D. | | Deposit date: | 1995-11-24 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and activity of a chimeric interleukin-8-melanoma-growth-stimulatory-activity protein.
Eur.J.Biochem., 235, 1996
|
|
5OPB
 
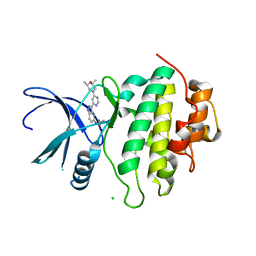 | | Structure of CHK1 10-pt. mutant complex with indazole LRRK2 inhibitor | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
2D7H
 
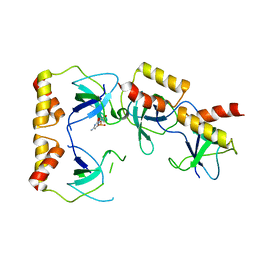 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
4M4R
 
 | | Epha4 ectodomain complex with ephrin a5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 4, ... | | Authors: | Xu, K, Tsvetkova-Robev, D, Xu, Y, Goldgur, Y, Chan, Y.-P, Himanen, J.P, Nikolov, D.B. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2D2Z
 
 | | Crystal structure of Soluble Form Of CLIC4 | | Descriptor: | Chloride intracellular channel protein 4 | | Authors: | Li, Y.F, Li, D.F, Wang, D.C. | | Deposit date: | 2005-09-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric structure of the wild soluble chloride intracellular ion channel CLIC4 observed in crystals
Biochem.Biophys.Res.Commun., 343, 2006
|
|
6EDB
 
 | | Crystal structure of SRY.hcGAS-21bp dsDNA complex | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*GP*GP*GP*AP*TP*CP*TP*AP*AP*AP*CP*AP*AP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*TP*GP*TP*TP*TP*AP*GP*AP*TP*CP*CP*CP*GP*GP*AP*TP*C)-3'), Sex-determining region Y protein,Cyclic GMP-AMP synthase, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5OY4
 
 | | GSK3beta complex with N-(6-(3,4-dihydroxyphenyl)-1H-pyrazolo[3,4-b]pyridin-3-yl)acetamide | | Descriptor: | Glycogen synthase kinase-3 beta, Proto-oncogene FRAT1, SULFATE ION, ... | | Authors: | Bax, B.D, Convery, M.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | From PIM1 to PI3K delta via GSK3 beta : Target Hopping through the Kinome.
ACS Med Chem Lett, 8, 2017
|
|
6QGB
 
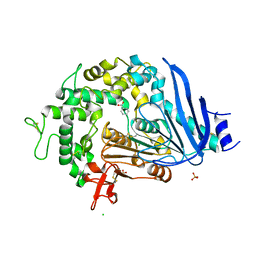 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
7RTI
 
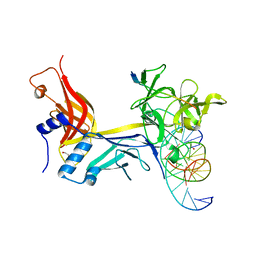 | | X-ray structure of RBPJ-L3MBTL3(dT62)-DNA complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Hall, D.P, Kovall, R.A, Yuan, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.
Nucleic Acids Res., 50, 2022
|
|
2OQ4
 
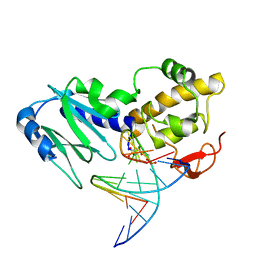 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli (E2Q) in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(*C*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*G*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Grollman, A.P, Shoahm, G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrates and mutations of critical residues
To be Published
|
|
307D
 
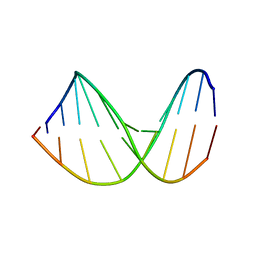 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
8QLR
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR24 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-2-[1,3-bis(oxidanyl)propan-2-ylamino]-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QLT
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR30 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[3-(2-oxidanylidenepyrrolidin-1-yl)propylamino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QLS
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR26 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(3-morpholin-4-ylpropylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6QFS
 
 | | Chargeless variant of the Cellulose-binding domain from Cellulomonas fimi | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Young, D.R, Hoejgaard, C, Messens, J, Winther, J.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Charge Interactions in a Highly Charge-depleted Protein
J.Am.Chem.Soc., 2021
|
|
3N92
 
 | | Crystal structure of TK1436, a GH57 branching enzyme from hyperthermophilic archaeon Thermococcus kodakaraensis, in complex with glucose | | Descriptor: | alpha-amylase, GH57 family, beta-D-glucopyranose | | Authors: | Santos, C.R, Tonoli, C.C.C, Trindade, D.M, Betzel, C, Takata, H, Kuriki, T, Kanai, T, Imanaka, T, Arni, R.K, Murakami, M.T. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus Kodakaraensis KOD1.
Proteins, 79, 2011
|
|
1UGC
 
 | |
6ZGF
 
 | | Spike Protein of RaTG13 Bat Coronavirus in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5XHW
 
 | | Crystal structure of HddC from Yersinia pseudotuberculosis | | Descriptor: | Putative 6-deoxy-D-mannoheptose pathway protein, SULFATE ION | | Authors: | Park, J, Kim, H, Kim, S, Shin, D.H. | | Deposit date: | 2017-04-24 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d-glycero-alpha-d-manno-heptose-1-phosphate guanylyltransferase from Yersinia pseudotuberculosis.
Biochim. Biophys. Acta, 1866, 2018
|
|
6Q38
 
 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
3NH9
 
 | | Nucleotide Binding Domain of Human ABCB6 (ATP bound structure) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family B member 6, mitochondrial, ... | | Authors: | Haffke, M, Menzel, A, Carius, Y, Jahn, D, Heinz, D.W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the nucleotide-binding domain of the human ABCB6 transporter and its complexes with nucleotides.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6Q4Q
 
 | | The Crystal structure of CK2a bound to P2-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, ACETATE ION, BENZOIC ACID, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
8QW7
 
 | | Crystal Structure of compound 4 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
5OP2
 
 | | Structure of CHK1 10-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
