1SYS
 
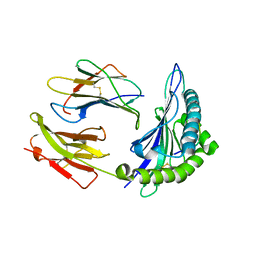 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
4RF2
 
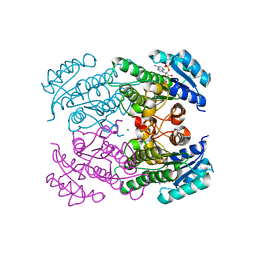 | |
5K9Y
 
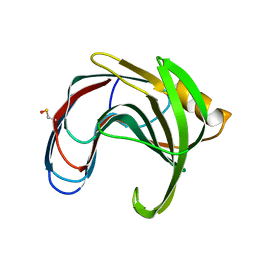 | | Crystal structure of a thermophilic xylanase A from Bacillus subtilis 1A1 quadruple mutant Q7H/G13R/S22P/S179C | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Pinheiro, M.P, Ferreira, T.L, Silva, S.R.B, Fuzo, C.A, Silva, S.R, Lourenzoni, M.R, Vieira, D.S, Ward, R.J, Nonato, M.C. | | Deposit date: | 2016-06-01 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of local residue environmental changes in thermostable mutants of the GH11 xylanase from Bacillus subtilis.
Int. J. Biol. Macromol., 97, 2017
|
|
1T3U
 
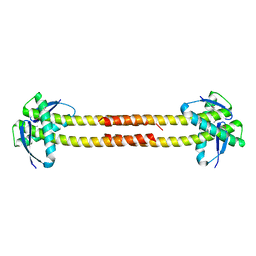 | | Unknown conserved bacterial protein from Pseudomonas aeruginosa PAO1 | | Descriptor: | conserved hypothetical protein | | Authors: | Rajashankar, K.R, Kneiwel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-04 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a conserved hypothetical protein Pseudomonas aeruginosa PA01
To be Published
|
|
1T7A
 
 | | Crystal structure of mutant Lys8Asp of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
5KGI
 
 | |
8A65
 
 | | Small molecule stabilizer (compound 3) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[2-azanyl-3,5-bis(chloranyl)phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
1TG3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, SULFATE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-28 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
7STB
 
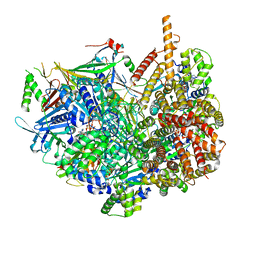 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ST9
 
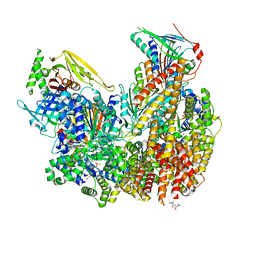 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5KGD
 
 | |
5K5F
 
 | | NMR structure of the HLTF HIRAN domain | | Descriptor: | Helicase-like transcription factor | | Authors: | Bezsonova, I, Neculai, D, Korzhnev, D, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HLTF HIRAN domain
To Be Published
|
|
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
7B5G
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
4RH8
 
 | |
4REV
 
 | | Structure of the dirigent protein DRR206 | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CHLORIDE ION, Disease resistance response protein 206, ... | | Authors: | Kim, K.-Y, Smith, C.A, Merkley, E.D, Cort, J.R, Davin, L.B, Lewis, N.G. | | Deposit date: | 2014-09-24 | | Release date: | 2014-11-26 | | Last modified: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Trimeric Structure of (+)-Pinoresinol-forming Dirigent Protein at 1.95 angstrom Resolution with Three Isolated Active Sites.
J.Biol.Chem., 290, 2015
|
|
4RF4
 
 | |
7T9Z
 
 | | Human Ornithine Aminotransferase (hOAT) crystallized at pH 6.0 | | Descriptor: | Ornithine aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Butrin, A, Liu, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Determination of the pH dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase.
J.Biol.Chem., 298, 2022
|
|
7TA0
 
 | | Human Ornithine Aminotransferase (hOAT) soaked with 5-aminovaleric acid | | Descriptor: | 5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Liu, D. | | Deposit date: | 2021-12-20 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Determination of the pH dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase.
J.Biol.Chem., 298, 2022
|
|
3ER9
 
 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with UU and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-R(UP*U)-3', CALCIUM ION, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
7ZRF
 
 | |
1TBU
 
 | |
2VNR
 
 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens. | | Descriptor: | CALCIUM ION, CPE0329 | | Authors: | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | Deposit date: | 2008-02-06 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
4RHJ
 
 | |
5KO4
 
 | | Bromodomain from Trypanosoma brucei Tb427.10.8150 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | El Bakkouri, M, Walker, J.R, Hou, C.F.D, Lin, Y.H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Bromodomain from Trypanosoma brucei Tb427.10.8150
To be published
|
|
