1TFZ
 
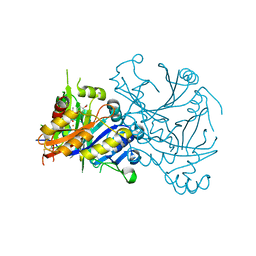 | | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
1JWW
 
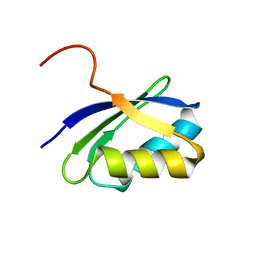 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
5KDL
 
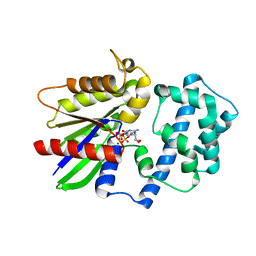 | | Crystal structure of the 4 alanine insertion variant of the Gi alpha1 subunit bound to GTPgammaS | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.665 Å) | | Cite: | A Conserved Hydrophobic Core in G alpha i1 Regulates G Protein Activation and Release from Activated Receptor.
J.Biol.Chem., 291, 2016
|
|
4IEB
 
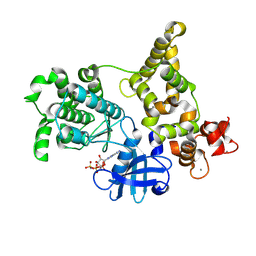 | | Crystal Structure of a Gly128Met mutant of the toxoplasma CDPK, TGME49_101440 | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wernimont, A.K, Artz, J.D, El Bakkouri, M, Schapira, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-13 | | Release date: | 2013-12-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a Gly128Met mutant of the toxoplasma CDPK, TGME49_101440
To be Published
|
|
3EUD
 
 | | Structure of the CS domain of the essential H/ACA RNP assembly protein Shq1p | | Descriptor: | Protein SHQ1 | | Authors: | Singh, M, Cascio, D, Gonzales, F.A, Heckmann, N, Chanfreau, G, Feigon, J. | | Deposit date: | 2008-10-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Functional Studies of the CS Domain of the Essential H/ACA Ribonucleoparticle Assembly Protein SHQ1.
J.Biol.Chem., 284, 2009
|
|
1K02
 
 | | Crystal Structure of Old Yellow Enzyme Mutant Gln114Asn | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, NADPH DEHYDROGENASE 1 | | Authors: | Brown, B.J, Hyun, J, Duvvuri, S.D, Karplus, P.A, Massey, V. | | Deposit date: | 2001-09-18 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of glutamine 114 in old yellow enzyme
J.Biol.Chem., 277, 2002
|
|
3EUV
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
4R8E
 
 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Yersinia pestis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Himiari, Z, Nanson, J.D, Swarbrick, C.M.D, Forwood, J.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Characterisation of the Beta-Ketoacyl-Acyl Carrier Protein Synthases, FabF and FabH, of Yersinia pestis.
Sci Rep, 5, 2015
|
|
1JTE
 
 | | Crystal Structure Analysis of VP39 F180W mutant | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1SXJ
 
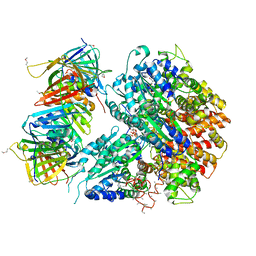 | | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Activator 1 37 kDa subunit, Activator 1 40 kDa subunit, ... | | Authors: | Bowman, G.D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Nature, 429, 2004
|
|
2IRV
 
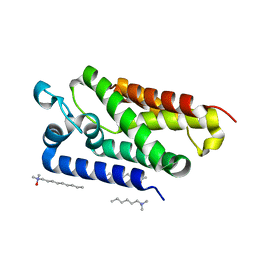 | | Crystal structure of GlpG, a rhomboid intramembrane serine protease | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, DODECYL-BETA-D-MALTOSIDE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Bibi, E, Fass, D, Ben-Shem, A. | | Deposit date: | 2006-10-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for intramembrane proteolysis by rhomboid serine proteases.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6IKG
 
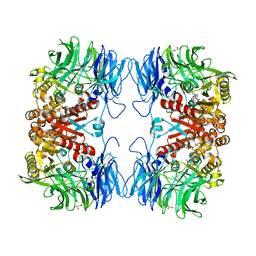 | | Crystal structure of substrate-bound S9 peptidase (S514A mutant) from Deinococcus radiodurans | | Descriptor: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-10-16 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
2ITF
 
 | | Crystal structure IsdA NEAT domain from Staphylococcus aureus with heme bound | | Descriptor: | Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
6W2E
 
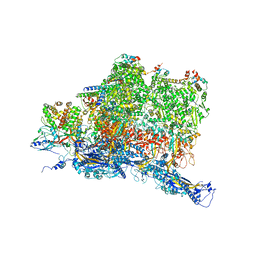 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
2IUR
 
 | |
3ZKV
 
 | |
1T2F
 
 | | Human B lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase B chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
5KA6
 
 | |
5K9B
 
 | | Azotobacter vinelandii Flavodoxin II | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin-2, MAGNESIUM ION | | Authors: | Rees, D.C, Segal, H.M, Spatzal, T. | | Deposit date: | 2016-05-31 | | Release date: | 2017-08-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.174 Å) | | Cite: | Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II.
Protein Sci., 26, 2017
|
|
1JR8
 
 | | Crystal Structure of Erv2p | | Descriptor: | Erv2 PROTEIN, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Gross, E, Sevier, C.S, Vala, A, Kaiser, C.A, Fass, D. | | Deposit date: | 2001-08-13 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p.
Nat.Struct.Biol., 9, 2002
|
|
4RC1
 
 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii with PRPP | | Descriptor: | PHOSPHATE ION, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E.J, Cascio, D, Sawaya, M.R, Yeates, T.O, Rasche, M.E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
3EKL
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
2J4E
 
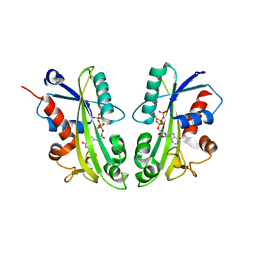 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | Descriptor: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | Authors: | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-08-29 | | Release date: | 2006-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
3EIE
 
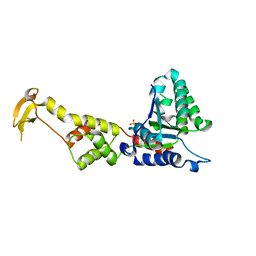 | | Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state | | Descriptor: | SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
