7B4T
 
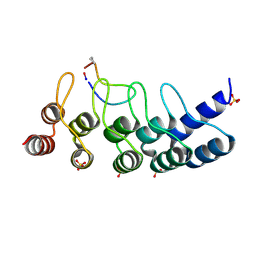 | | Broadly neutralizing DARPin bnD.1 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | Broadly neutralizing DARPin bnD.1, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), SULFATE ION | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
4V8L
 
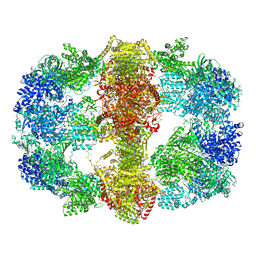 | |
7B4W
 
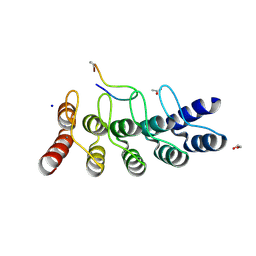 | | Broadly neutralizing DARPin bnD.3 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Broadly neutralizing DARPin bnD.3, ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
3E6F
 
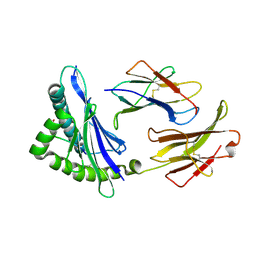 | | MHC CLASS I H-2Dd Heavy chain complexed with Beta-2 Microglobulin and a variant peptide, PA9, from the Human immunodeficiency virus (BaL) envelope glycoprotein 120 | | Descriptor: | BETA-2 MICROGLOBULIN, Envelope glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Wang, R, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
J.Immunol., 183, 2009
|
|
5K6I
 
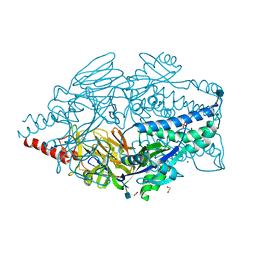 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 A149C-Y458C, S46G-E92D-S215P-K465Q variant. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1SZS
 
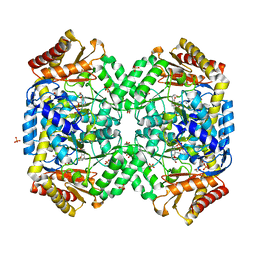 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
6W1G
 
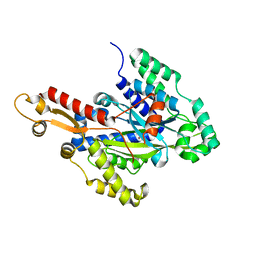 | | Crystal structure of the hydroxyglutarate synthase from Pseudomonas putida | | Descriptor: | Hydroxyglutarate synthase, NICKEL (II) ION | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
5K7N
 
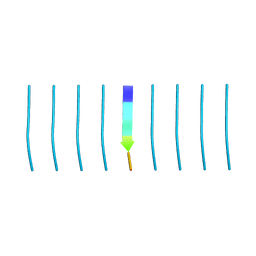 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
7B2N
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic Fructose bisphosphate aldolase | | Descriptor: | CHLORIDE ION, Fructose-bisphosphate aldolase 1, chloroplastic, ... | | Authors: | Le Moigne, T, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of chloroplast fructose-1,6-bisphosphate aldolase from the green alga Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2022
|
|
2J14
 
 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4UYR
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
1TC0
 
 | | Ligand Induced Conformational Shifts in the N-terminal Domain of GRP94, Open Conformation Complexed with the physiological partner ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
6WAH
 
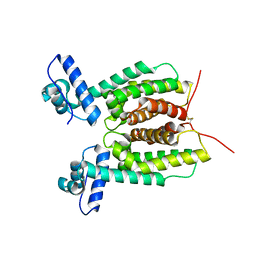 | | Crystal Structure of SmcR L139R from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
4V6D
 
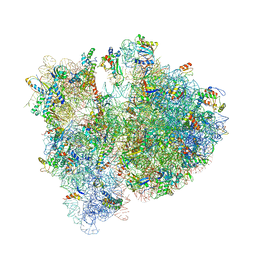 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7U
 
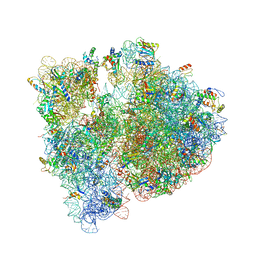 | | Crystal structure of the E. coli ribosome bound to erythromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8V
 
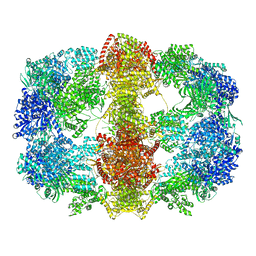 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
108D
 
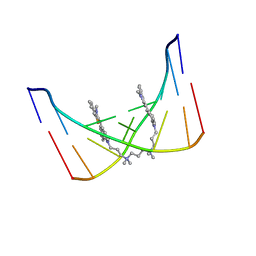 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
1TA1
 
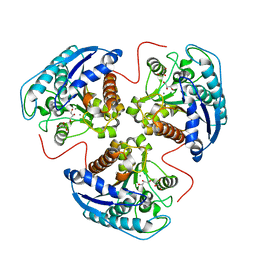 | | H141C mutant of rat liver arginase I | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-19 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
5KGF
 
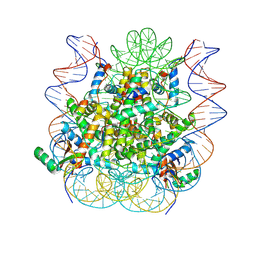 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
4UWG
 
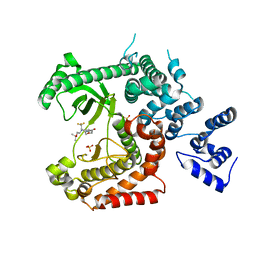 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
1JRG
 
 | | Crystal Structure of the R3 form of Pectate Lyase A, Erwinia chrysanthemi | | Descriptor: | Pectate lyase, SULFATE ION | | Authors: | Thomas, L.M, Doan, C, Oliver, R.L, Yoder, M.D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pectate lyase A: comparison to other isoforms.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
6PZN
 
 | |
