7A6J
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Poziotinib | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
8OFA
 
 | | Crystal structure of human cathepsin L interacting with tosyl phenylalanyl chloromethyl ketone (TPCK) | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 4-methyl-~{N}-[(2~{S})-4-oxidanyl-3-oxidanylidene-1-phenyl-butan-2-yl]benzenesulfonamide, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Ewert, W, Reinke, P.Y.A, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-03-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
4XQ8
 
 | |
5FWQ
 
 | | Apo structure of human Leukotriene A4 hydrolase | | Descriptor: | ACETATE ION, HUMAN LEUKOTRIENE A4 HYDROLASE, IMIDAZOLE, ... | | Authors: | Wittmann, S.K, Kalinowsky, L, Kramer, J, Bloecher, R, Steinhilber, D, Pogoryelov, D, Proschak, E, Heering, J. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Thermodynamic properties of leukotriene A4hydrolase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
8SB6
 
 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
8K0B
 
 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | PYROGLUTAMYL PEPTIDASE-1 | | Authors: | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | Deposit date: | 1997-08-26 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
5JLT
 
 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
6RRO
 
 | |
3LP9
 
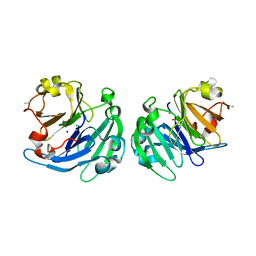 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
6RRL
 
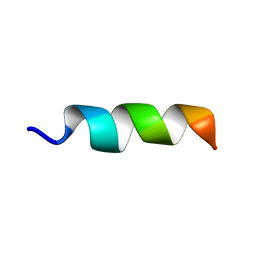 | |
1B7T
 
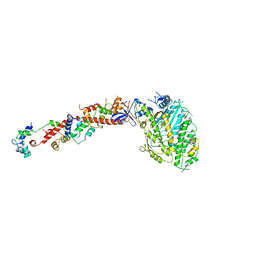 | | MYOSIN DIGESTED BY PAPAIN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Houdusse, A, Kalabokis, V, Himmel, D, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head.
Cell(Cambridge,Mass.), 97, 1999
|
|
5MBB
 
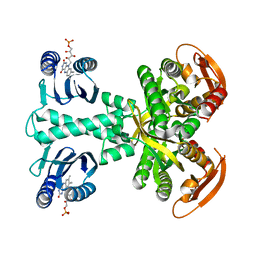 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
3LEX
 
 | | 2F5 Epitope scaffold elicited anti-HIV-1 monoclonal antibody 11F10 in complex with HIV-1 GP41 | | Descriptor: | 11f10 Antibody Heavy Chain, 11f10 Antibody Light Chain, Envelope glycoprotein gp41 | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Baker, D, Wyatt, R, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8S93
 
 | | Crystal structure of the PH-TH/kinase complex of Bruton's tyrosine kinase | | Descriptor: | 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, GLYCEROL, Tyrosine-protein kinase BTK, ... | | Authors: | Lin, D.Y, Andreotti, A.H. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational heterogeneity of the BTK PHTH domain drives multiple regulatory states.
Elife, 12, 2024
|
|
5U6E
 
 | | Crystal structure of clade A/E HIV-1 gp120 core in complex with NBD-14010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{(1S)-2-amino-1-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl]ethyl}-5-(4-chloro-3-fluorophenyl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
J. Med. Chem., 60, 2017
|
|
5MP7
 
 | | Crystal structure of phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | Donini, S, Garavaglia, S, Ferraris, D.M, Miggiano, R, Mori, S, Shibayama, K, Rizzi, M. | | Deposit date: | 2016-12-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural investigations on phosphoribosylpyrophosphate synthetase from Mycobacterium smegmatis.
PLoS ONE, 12, 2017
|
|
6M7G
 
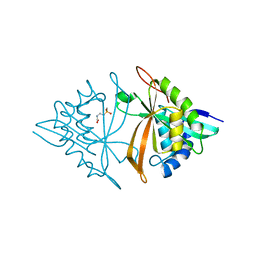 | | Crystal structure of ArsN, N-acetyltransferase with substrate phosphinothricin from Pseudomonas putida KT2440 | | Descriptor: | PHOSPHINOTHRICIN, Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
8G4L
 
 | | Cryo-EM structure of the human cardiac myosin filament | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | Dutta, D, Nguyen, V, Padron, R, Craig, R. | | Deposit date: | 2023-02-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-EM structure of the human cardiac myosin filament.
Nature, 623, 2023
|
|
5FYR
 
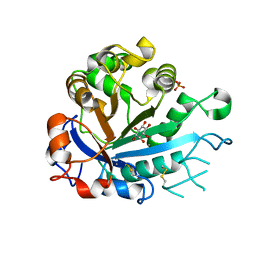 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, myoinositol complex | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MQL
 
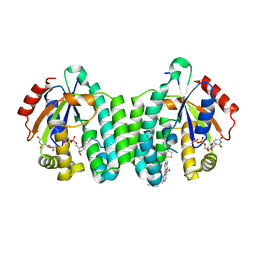 | | Crystal structure of dCK mutant C3S in complex with masitinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, Deoxycytidine kinase, MAGNESIUM ION, ... | | Authors: | Rebuffet, E, Hammam, K, Saez-Ayala, M, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
7JN3
 
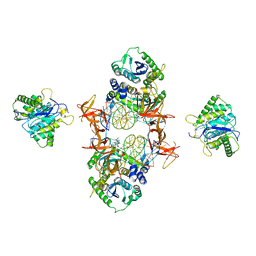 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
