1KQN
 
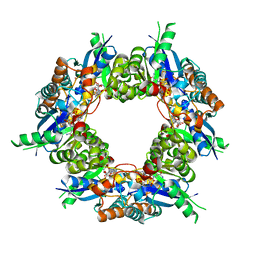 | | Crystal structure of NMN/NaMN adenylyltransferase complexed with NAD | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XENON | | Authors: | Zhou, T, Kurnasov, O, Tomchick, D.R, Binns, D.D, Grishin, N.V, Marquez, V.E, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-01-07 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human Nicotinamide/Nicotonic Acid Mononucleotide Adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin.
J.Biol.Chem., 277, 2003
|
|
6EJ7
 
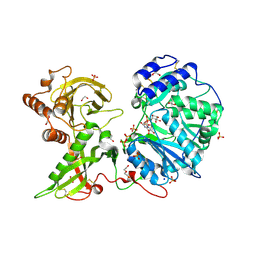 | |
1KFE
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH L-Ser Bound To The Beta Site | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1UGD
 
 | |
6QND
 
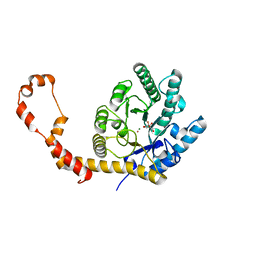 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 60 s timepoint | | Descriptor: | COBALT (II) ION, D-glucose, MAGNESIUM ION, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-11 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
2OPP
 
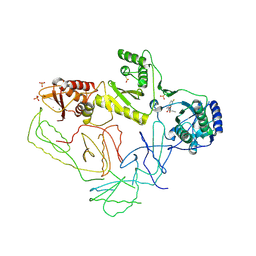 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
6QPH
 
 | | Dunaliella minimal PSI complex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2019-02-14 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
3NIQ
 
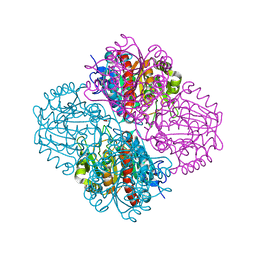 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase | | Descriptor: | 3-guanidinopropionase, GLYCEROL, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
8QV0
 
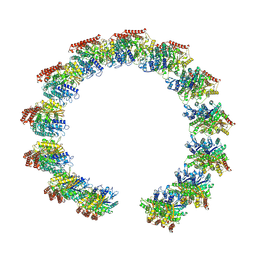 | | Structure of the native microtubule lattice nucleated from the yeast spindle pole body | | Descriptor: | Tubulin alpha-1 chain, Tubulin beta chain | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4LSS
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC01 in complex with HIV-1 clade A strain KER_2018_11 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY VRC01, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
1KFC
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM With Indole Propanol Phosphate | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
3NK1
 
 | | Complex of 6-hydroxy-L-nicotine oxidase with serotonin | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2010-06-18 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure analysis of free and substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
4MI6
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[4-(5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[4-(5,6,7,8-tetrahydronaphthalen-2-yl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
3NMD
 
 | | Crystal structure of the leucine zipper domain of cGMP dependent protein kinase I beta | | Descriptor: | GLYCEROL, HEXANE-1,6-DIOL, cGMP Dependent PRotein Kinase | | Authors: | Kim, C, Casteel, D.E, Smith-Nguyen, E.V, Sankaran, B, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring.
J.Biol.Chem., 285, 2010
|
|
6QZK
 
 | | Structure of Clostridium butyricum Argonaute bound to a guide DNA (5' deoxycytidine) and a 19-mer target DNA | | Descriptor: | Clostridium butyricum Argonaute, DNA target (5'-D(T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*T)-3'), FORMIC ACID, ... | | Authors: | Swarts, D.C, Jinek, M, Hegge, J.W, Van der Oost, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.548 Å) | | Cite: | DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Nucleic Acids Res., 47, 2019
|
|
1TWF
 
 | | RNA polymerase II complexed with UTP at 2.3 A resolution | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
8Q7E
 
 | |
8QV3
 
 | | Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, Spindle pole body component 110, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3NEX
 
 | |
4M3Q
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3NIE
 
 | | Crystal Structure of PF11_0147 | | Descriptor: | MAP2 kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Cossar, D, Amani, M, Artz, J.D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of PF11_0147
To be Published
|
|
5BQS
 
 | | S. Pneumoniae Fabh with small molecule inhibitor 4 | | Descriptor: | 1-{5-[2-chloro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SODIUM ION | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-29 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
1KGW
 
 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337Q VARIANT OF HUMAN PANCREATIC ALPHA-MYLASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
4MGR
 
 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MHS
 
 | | Crystal structure of Gpb in complex with SUGAR (N-[(2E)-3-(BIPHENYL-4-YL)PROP-2-ENOYL]-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
