5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | 著者 | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | 登録日 | 2016-06-16 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | 分子名称: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | 著者 | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | 登録日 | 2019-04-02 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.517 Å) | | 主引用文献 | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
6DMH
 
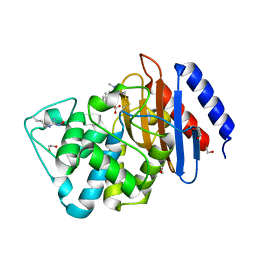 | | A multiconformer ligand model of acylenzyme intermediate of meropenem bound to an SFC-1 E166A mutant | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | 著者 | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | 登録日 | 2018-06-05 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
6DN0
 
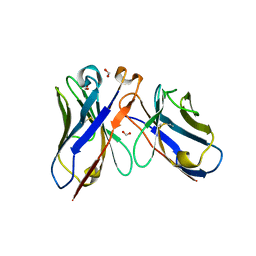 | |
8RXR
 
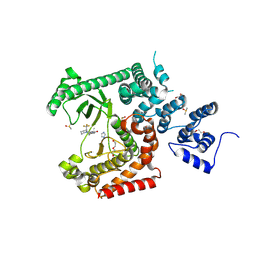 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | 分子名称: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | 登録日 | 2024-02-07 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-08-21 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
4V1X
 
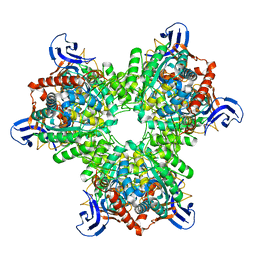 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | 分子名称: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | 著者 | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | 登録日 | 2014-10-04 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WFS
 
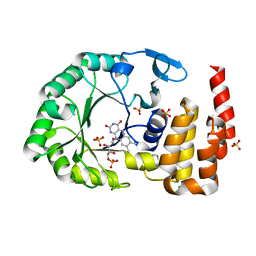 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | 分子名称: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | 著者 | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | 登録日 | 2014-09-17 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
2G5Z
 
 | | Structure of S65G Y66S GFP variant after spontaneous peptide hydrolysis and decarboxylation | | 分子名称: | Green fluorescent protein, MAGNESIUM ION | | 著者 | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2006-02-23 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Understanding GFP Posttranslational Chemistry: Structures of Designed Variants that Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation.
J.Am.Chem.Soc., 128, 2006
|
|
4WHB
 
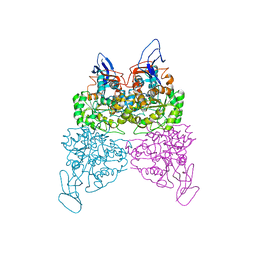 | | Crystal structure of phenylurea hydrolase B | | 分子名称: | Phenylurea hydrolase B, ZINC ION | | 著者 | Sugrue, E, Carr, P.D, Khurana, J.L, Jackson, C.J. | | 登録日 | 2014-09-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.958 Å) | | 主引用文献 | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
6E15
 
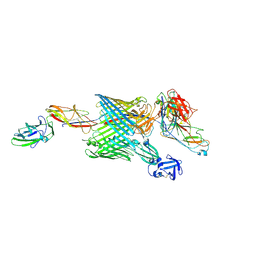 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | 分子名称: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | 著者 | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | 登録日 | 2018-07-09 | | 公開日 | 2018-10-17 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
6E1W
 
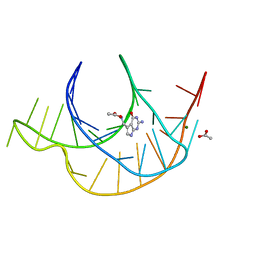 | | Crystal structure of a class I PreQ1 riboswitch complexed with PreQ1 | | 分子名称: | 2-amino-5-(aminomethyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | 登録日 | 2018-07-10 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1S
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | 分子名称: | 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, RNA (33-MER) | | 著者 | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | 登録日 | 2018-07-10 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | 分子名称: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | 著者 | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2014-10-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
4WPY
 
 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-II) | | 分子名称: | GLYCEROL, protein DL-Rv1738, trifluoroacetic acid | | 著者 | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | 登録日 | 2014-10-21 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WSC
 
 | | Crystal structure of a GroELK105A mutant | | 分子名称: | 60 kDa chaperonin | | 著者 | Lorimer, G.H, Ye, X, Fei, X, Yang, D, Corsepius, N, LaRonde, N.A. | | 登録日 | 2014-10-26 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Crystal structure of a GroELK105A mutant
To Be Published
|
|
4WKS
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | Acyl-homoserine lactone acylase PvdQ, ethylboronic acid | | 著者 | Wu, R, Clevenger, D.K, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
7RT5
 
 | | Crystal structure of KRAS G12D with compound 36 (4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine) bound | | 分子名称: | 4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | 登録日 | 2021-08-12 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT3
 
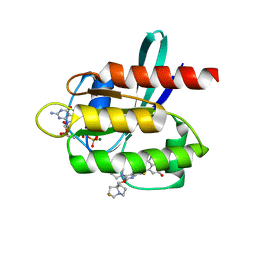 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | 分子名称: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | 著者 | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | 登録日 | 2021-08-12 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT4
 
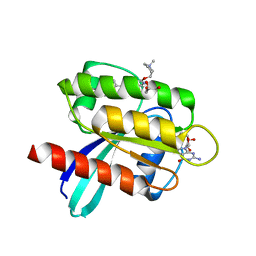 | | KRAS G12D in complex with Compound 5B (7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine) | | 分子名称: | 7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | 登録日 | 2021-08-12 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7S97
 
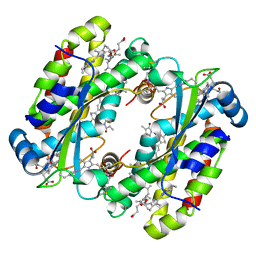 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | 分子名称: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | 著者 | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | 登録日 | 2021-09-20 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7D30
 
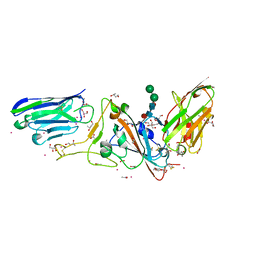 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | 分子名称: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | 著者 | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | 登録日 | 2020-09-17 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
4USX
 
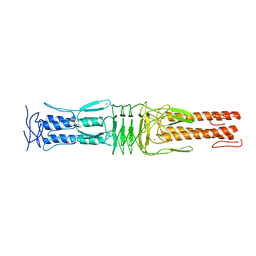 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | 分子名称: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | 著者 | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6DTN
 
 | |
4UUM
 
 | |
5DHX
 
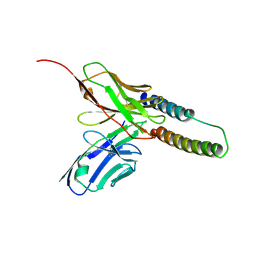 | | HIV-1 Rev NTD dimers with variable crossing angles | | 分子名称: | Anti-Rev Antibody Fab single-chain variable fragment, light chain,Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, ... | | 著者 | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | 登録日 | 2015-08-31 | | 公開日 | 2016-06-22 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
