8DAU
 
 | |
8DAR
 
 | |
8TSO
 
 | | KDL bound, nucleotide-free MsbA in open, outward-facing conformation | | 分子名称: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ATP-binding transport protein MsbA, PENTAETHYLENE GLYCOL MONODECYL ETHER | | 著者 | Yang, B, Zhang, T, Lyu, J, Laganowsky, A.D, Zhao, M. | | 登録日 | 2023-08-11 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Native mass spectrometry captures snapshots of the MsbA transport cycle
To Be Published
|
|
8DAS
 
 | |
5MC9
 
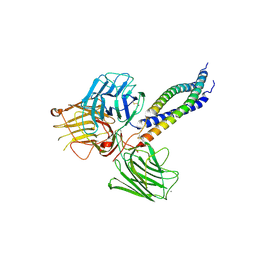 | |
8DAT
 
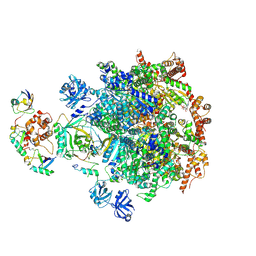 | |
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | 分子名称: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
8DAV
 
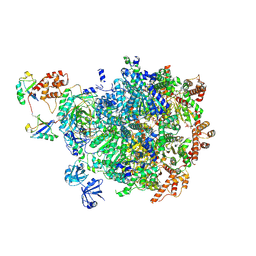 | |
5A2K
 
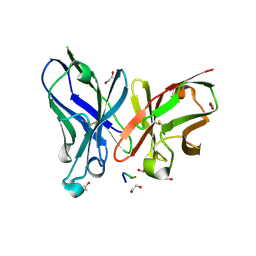 | | Crystal structure of scFv-SM3 in complex with APD-TGalNAc-RP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | 著者 | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | 登録日 | 2015-05-20 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6C2R
 
 | | Aurora A ligand complex | | 分子名称: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | 著者 | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | 登録日 | 2018-01-08 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6C2Z
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | 分子名称: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | 著者 | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | 登録日 | 2018-01-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
5MG7
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-p53R2) | | 分子名称: | Cellular tumor antigen p53, DNA, ZINC ION | | 著者 | Rozenberg, H, Braeuning, B, Golovenko, D, Shakked, Z. | | 登録日 | 2016-11-21 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
8D38
 
 | | Structure of a purine nucleoside phosphorylase from Geobacillus stearothermophilus | | 分子名称: | Purine nucleoside phosphorylase, SODIUM ION | | 著者 | Given, F, Johnston, J, Crittenden, D, Moran, F, Johns, A. | | 登録日 | 2022-05-31 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | The structure of His-tagged Geobacillus stearothermophilus purine nucleoside phosphorylase reveals a 'spanner in the works'.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6FHL
 
 | | Cryo-EM structure of F-actin in complex with ADP-Pi | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Merino, F, Pospich, S, Funk, J, Wagner, T, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | 登録日 | 2018-01-15 | | 公開日 | 2018-06-13 | | 最終更新日 | 2018-08-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8U57
 
 | | PPARg LBD in complex with perfluorooctanoic acid (PFOA) | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Peroxisome proliferator-activated receptor gamma, pentadecafluorooctanoic acid | | 著者 | Pederick, J.L, Frkic, R.L, McDougal, D.P, Bruning, J.B. | | 登録日 | 2023-09-12 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A structural basis for the activation of peroxisome proliferator-activated receptor gamma (PPAR gamma ) by perfluorooctanoic acid (PFOA).
Chemosphere, 354, 2024
|
|
6FI0
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with Fr 19 | | 分子名称: | 2-azanyl-1-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl)ethanone, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | 著者 | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | 登録日 | 2018-01-16 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6T1A
 
 | | Structure of mosquitocidal Cyt1Aa protoxin obtained by Serial Femtosecond Crystallography on in vivo grown crystals at pH 10 | | 分子名称: | CALCIUM ION, Type-1Aa cytolytic delta-endotoxin | | 著者 | Tetreau, G, Banneville, A.S, Andreeva, E, Brewster, A.S, Hunter, M.S, Sierra, R.G, Young, I.D, Boutet, S, Coquelle, N, Cascio, D, Sawaya, M.R, Sauter, N.K, Colletier, J.P. | | 登録日 | 2019-10-03 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade.
Nat Commun, 11, 2020
|
|
6VSI
 
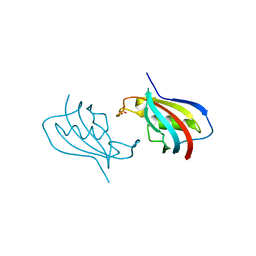 | | Crystal structure of FKBP12 of Candida auris | | 分子名称: | Peptidylprolyl isomerase, SULFATE ION | | 著者 | Li, Z, Li, H, Hernandez, G, LeMaster, D. | | 登録日 | 2020-02-11 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure and transient dimerization for the FKBP12 protein from the pathogenic fungus Candida auris.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
5AH2
 
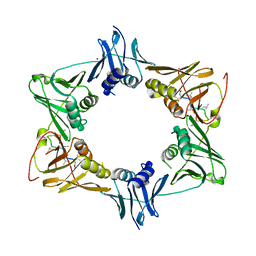 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | 分子名称: | DNA POLYMERASE III SUBUNIT BETA, GRISELIMYCIN, SODIUM ION | | 著者 | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | 登録日 | 2015-02-04 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.129 Å) | | 主引用文献 | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
6T1F
 
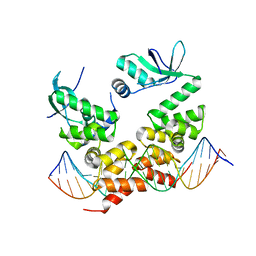 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed to the centromeric parS site | | 分子名称: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*CP*C)-3') | | 著者 | Jalal, A.S.B, Pastrana, C.L, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Moreno-Herrero, F, Le, T.B.K. | | 登録日 | 2019-10-04 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
6BM9
 
 | |
8TE7
 
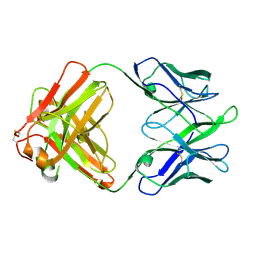 | | Structure of TRNM-f.01 | | 分子名称: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | 著者 | Bender, M.F, Olia, A.S, Kwong, P.D. | | 登録日 | 2023-07-05 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Broad and Potent HIV-1 Neutralization in Fusion Peptide-primed SHIV-boosted Macaques
To Be Published
|
|
8D4O
 
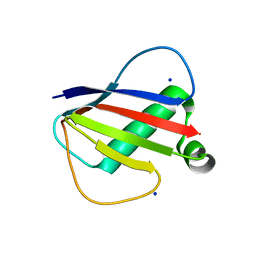 | |
4WR2
 
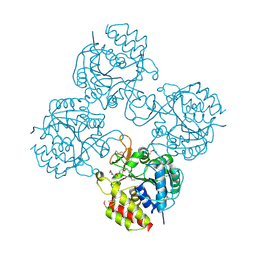 | | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site | | 分子名称: | CALCIUM ION, PENTAETHYLENE GLYCOL, Pyrimidine-specific ribonucleoside hydrolase RihA | | 著者 | Himmel, D.M, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-10-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of a putative pyrimidine-specific ribonucleoside hydrolase (RihA) Protein from Shewanella loihica PV-4 (SHEW_0697, Target PSI-029635) with divalent cation and PEG 400 bound at the active site
To be published
|
|
