4Y63
 
 | | AAGlyB in complex with amino-acid analogues | | 分子名称: | 5'-deoxy-5'-{[(2S)-2-(triaza-1,2-dien-2-ium-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | 著者 | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | 登録日 | 2015-02-12 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
8IO8
 
 | |
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-21 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
8U3J
 
 | |
8PKU
 
 | | Kelch domain of KEAP1 in complex with ortho-dimethylbenzene linked cyclic peptide 3 (ortho-WRCDEETGEC). | | 分子名称: | (2-methylphenyl)methanol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Braun, M.B, Bischof, L, Hartmann, M.D. | | 登録日 | 2023-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Computational Prediction of Cyclic Peptide Structural Ensembles and Application to the Design of Keap1 Binders.
J.Chem.Inf.Model., 63, 2023
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | 分子名称: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2022-01-26 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6VK7
 
 | | Crystal Structure of reduced Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | 分子名称: | FE (III) ION, Methane monooxygenase, Methane monooxygenase component A alpha chain | | 著者 | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | 登録日 | 2020-01-18 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
5UK0
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
6GND
 
 | | Crystal structure of the complex of a Ferredoxin-Flavin Thioredoxin Reductase and a Thioredoxin from Clostridium acetobutylicum at 2.9 A resolution | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin, ... | | 著者 | Buey, R.M, Fernandez-Justel, D, Balsera, M. | | 登録日 | 2018-05-30 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.889 Å) | | 主引用文献 | Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | 分子名称: | Tetrahedral aminopeptidase, ZINC ION | | 著者 | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | 登録日 | 2019-04-02 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | 主引用文献 | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
3I3D
 
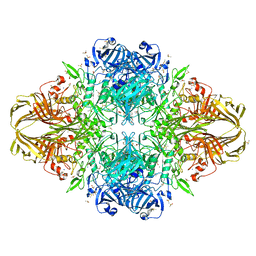 | | E. COLI (lacZ) BETA-GALACTOSIDASE (M542A) IN COMPLEX WITH IPTG | | 分子名称: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Dugdale, M.L, Dymianiw, D, Minhas, B, Huber, R.E. | | 登録日 | 2009-06-30 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Role of Met-542 as a guide for the conformational changes of Phe-601 that occur during the reaction of β-galactosidase (Escherichia coli).
Biochem.Cell Biol., 88, 2010
|
|
6WJH
 
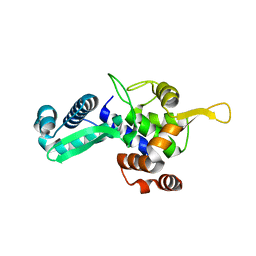 | |
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | 分子名称: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | 登録日 | 2022-01-26 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7ZNP
 
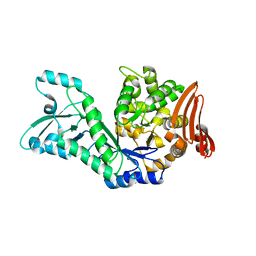 | |
6RBE
 
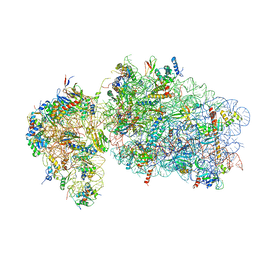 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
7TVD
 
 | |
5A3O
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(cinnamido)-6-deoxy-alpha-D-mannopyranoside at 1.6 ansgtrom | | 分子名称: | CALCIUM ION, CHLORIDE ION, CINNAMIDE, ... | | 著者 | Sommer, R, Hauck, D, Varrot, A, Audfray, A, Imberty, A, Titz, A. | | 登録日 | 2015-06-02 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Cinnamide Derivatives of D-Mannose as Inhibitors of the Bacterial Virulence Factor Lecb from Pseudomonas Aeruginosa
Chemistryopen, 4, 2015
|
|
8U3L
 
 | |
7ZPW
 
 | | Crystal structure of Pizza6-TSK-TSH with Silicotungstic Acid (STA) polyoxometalate | | 分子名称: | Keggin (STA), Pizza6-TSK-TSH | | 著者 | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | 登録日 | 2022-04-29 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mutational study of a symmetry matched protein-polyoxometalate interface.
To be published
|
|
5K31
 
 | | Crystal structure of Human fibrillar procollagen type I C-propeptide Homo-trimer | | 分子名称: | CALCIUM ION, CHLORIDE ION, Collagen alpha-1(I) chain, ... | | 著者 | Sharma, U, Hulmes, D.J.S, Aghajari, N. | | 登録日 | 2016-05-19 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of homo- and heterotrimerization of collagen I.
Nat Commun, 8, 2017
|
|
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-08-04 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
4Y62
 
 | | AAGlyB in complex with amino-acid analogues | | 分子名称: | 5'-deoxy-5'-{[(2R)-3-hydroxy-2-(4-phenyl-1H-1,2,3-triazol-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | 著者 | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | 登録日 | 2015-02-12 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
6W5H
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
8TBY
 
 | | Apo Bcs1, unsymmetrized | | 分子名称: | Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-06-29 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8I3Z
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | 著者 | Peng, X, Lilley, D.M.J, Huang, L. | | 登録日 | 2023-01-18 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
