7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7BJT
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | 分子名称: | Alginate lyase, family PL17, CALCIUM ION, ... | | 著者 | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | 登録日 | 2021-01-14 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
6NDG
 
 | |
6PTZ
 
 | | Crystal structure of pigeon Cryptochrome 4 mutant Y319D in complex with flavin adenine dinucleotide | | 分子名称: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | 登録日 | 2019-07-16 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FS0
 
 | |
6TTR
 
 | | Crystal Structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GGDEF diguanylate cyclase DgcB, PHOSPHATE ION | | 著者 | Holzschuh, F, Schirmer, T, Teixeira, R.D. | | 登録日 | 2019-12-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal structure of the coiled coil and GGDEF domain of DgcB from Caulobacter crescentus in complex with c-di-GMP
To Be Published
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | 分子名称: | Nsp9, SULFATE ION | | 著者 | Zhang, C, Chen, Y, Li, L, Su, D. | | 登録日 | 2020-04-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.954 Å) | | 主引用文献 | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
7LQW
 
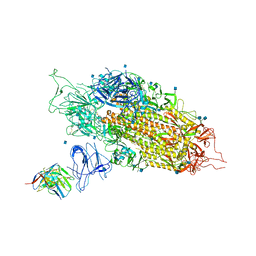 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | 分子名称: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (4.47 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
6NDA
 
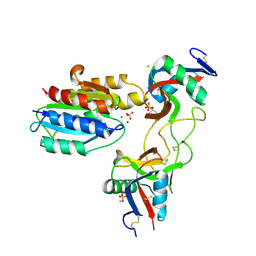 | |
6TH0
 
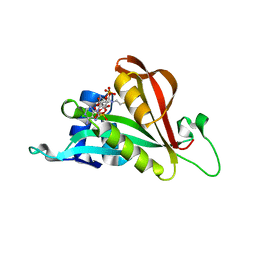 | | Crystal structure of Arabidopsis thaliana NAA60 in complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, Acyl-CoA N-acyltransferases (NAT) superfamily protein | | 著者 | Layer, D, Kopp, J, Lapouge, K, Sinning, I. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Arabidopsis N alpha -acetyltransferase NAA60 locates to the plasma membrane and is vital for the high salt stress response.
New Phytol., 228, 2020
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBN
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s3 [t2 dataset] | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | 著者 | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | 登録日 | 2021-08-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
5WY8
 
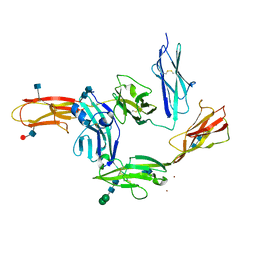 | | Crystal structure of PTP delta Ig1-Ig3 in complex with IL1RAPL1 Ig1-Ig3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | 著者 | Kim, H.M, Won, S.Y, Kim, D. | | 登録日 | 2017-01-11 | | 公開日 | 2017-11-22 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | LAR-RPTP Clustering Is Modulated by Competitive Binding between Synaptic Adhesion Partners and Heparan Sulfate
Front Mol Neurosci, 10, 2017
|
|
3JRB
 
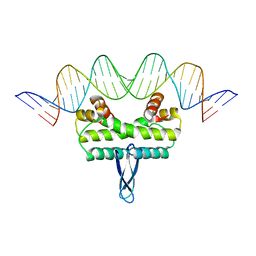 | |
8Q85
 
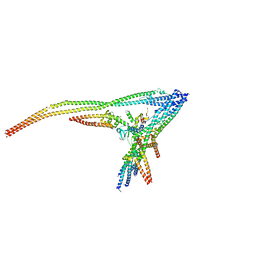 | |
3JRH
 
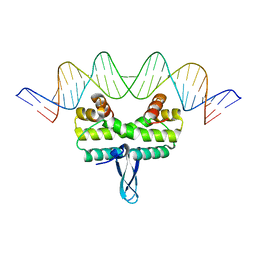 | |
6Q41
 
 | | Atomic resolution crystal structure of a BAA collagen heterotrimer | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Leading chain of the BAA collagen heterotrimer, ... | | 著者 | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
7S23
 
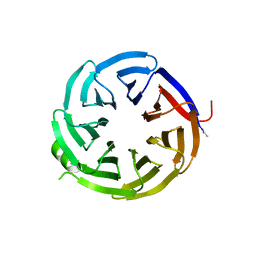 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | 分子名称: | Coatomer subunit alpha | | 著者 | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | 登録日 | 2021-09-03 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
8RDP
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | 分子名称: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | 著者 | Bischof, L, Hartmann, M.D. | | 登録日 | 2023-12-08 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | 分子名称: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | 分子名称: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | 分子名称: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | 分子名称: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
