7ZQ8
 
 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) | | 分子名称: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | 著者 | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | 登録日 | 2022-04-29 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
7ZJZ
 
 | | catalytically non active S532A mutant of oligopeptidase B from S. proteomaculans | | 分子名称: | Oligopeptidase B, SPERMINE | | 著者 | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | 登録日 | 2022-04-12 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
3EMN
 
 | | The Crystal Structure of Mouse VDAC1 at 2.3 A resolution | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | 著者 | Ujwal, R, Cascio, D, Colletier, J.-P, Faham, S, Zhang, J, Toro, L, Ping, P, Abramson, J. | | 登録日 | 2008-09-24 | | 公開日 | 2008-12-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6TCI
 
 | |
8FLQ
 
 | | Human PTH1R in complex with PTH(1-34) and Gs | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Cary, B.P, Belousoff, M.J, Piper, S.J, Wootten, D, Sexton, P.M. | | 登録日 | 2022-12-22 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Molecular insights into peptide agonist engagement with the PTH receptor.
Structure, 31, 2023
|
|
8FWN
 
 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | 登録日 | 2023-01-23 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
6WHE
 
 | |
3IKY
 
 | | Structural model of ParM filament in the open state by cryo-EM | | 分子名称: | Plasmid segregation protein parM | | 著者 | Galkin, V.E, Orlova, A, Rivera, C, Mullins, R.D, Egelman, E.H. | | 登録日 | 2009-08-06 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Structural polymorphism of the ParM filament and dynamic instability
Structure, 17, 2009
|
|
6TE2
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | 分子名称: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha' | | 著者 | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | 登録日 | 2019-11-11 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (0.922 Å) | | 主引用文献 | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
4GLI
 
 | |
6TE8
 
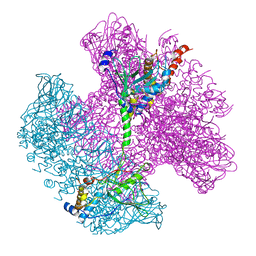 | | Neck of native GTA particle computed with C12 symmetry | | 分子名称: | Adaptor protein Rcc01688, Phage portal protein, HK97 family | | 著者 | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | 登録日 | 2019-11-11 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
5XFZ
 
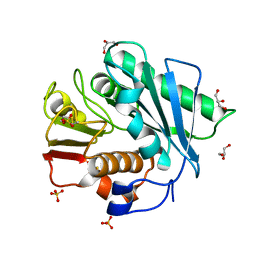 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | 分子名称: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | 著者 | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2017-04-11 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
6WZS
 
 | | Fusibacterium ulcerans ZTP riboswitch bound to m-1-pyridinyl AICA | | 分子名称: | 5-amino-1-(pyridin-3-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | 著者 | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | 登録日 | 2020-05-14 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
6TEB
 
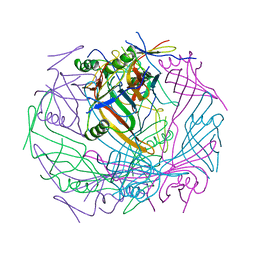 | | Tail-baseplate interface of native GTA particle computed with C6 symmetry | | 分子名称: | Distal tail protein Rcc01695, Tail tube protein Rcc01691 | | 著者 | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | 登録日 | 2019-11-11 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6WZR
 
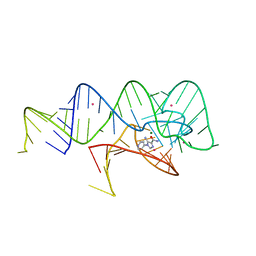 | | Fusibacterium ulcerans ZTP riboswitch bound to p-1-pyridinyl AICA | | 分子名称: | 5-amino-1-(pyridin-4-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | 著者 | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | 登録日 | 2020-05-14 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
8FO0
 
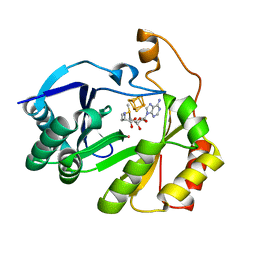 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | 分子名称: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | 著者 | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2022-12-29 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
6WX2
 
 | |
8FOL
 
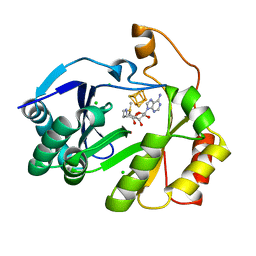 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2022-12-31 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
6NZZ
 
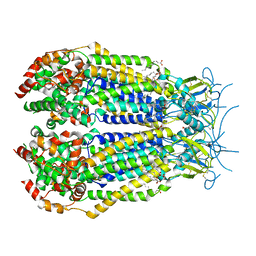 | | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}butanoic acid, Volume-regulated anion channel subunit LRRC8A | | 著者 | Kern, D.M, Hite, R.K, Brohawn, S.G. | | 登録日 | 2019-02-14 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs.
Elife, 8, 2019
|
|
6HHX
 
 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)cytidine | | 分子名称: | 5'-O-(N-(L-aspartyl)-sulfamoyl)cytidine, Aspartate--tRNA(Asp) ligase | | 著者 | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2018-08-29 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
5HZY
 
 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | 分子名称: | Uncharacterized protein RavZ | | 著者 | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | 登録日 | 2016-02-03 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.548 Å) | | 主引用文献 | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
3IS2
 
 | | 2.3 Angstrom Crystal Structure of a Cys71 Sulfenic Acid form of Vivid | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Vivid PAS protein VVD | | 著者 | Zoltowski, B.D, Lamb, J.S, Pabit, S.A, Li, L, Pollack, L, Crane, B.R. | | 登録日 | 2009-08-25 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Illuminating solution responses of a LOV domain protein with photocoupled small-angle X-ray scattering.
J.Mol.Biol., 393, 2009
|
|
6HHV
 
 | | Structure of T. thermophilus AspRS in Complex with 5'-O-(N-(L-aspartyl)-sulfamoyl)N3-methyluridine | | 分子名称: | 5'-O-(N-(L-aspartyl)-sulfamoyl)N3-methyluridine, Aspartate--tRNA(Asp) ligase | | 著者 | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2018-08-29 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
3IKD
 
 | | Structure-Based Design of Novel PIN1 Inhibitors (I) | | 分子名称: | (2R)-2-[(1-benzothiophen-2-ylcarbonyl)amino]-3-phenylpropyl phosphate, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Matthews, D, Greasley, S, Ferre, R, Parge, H. | | 登録日 | 2009-08-05 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of novel human Pin1 inhibitors (I).
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8FSI
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2023-01-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
