8CRS
 
 | |
5LDD
 
 | |
7AV7
 
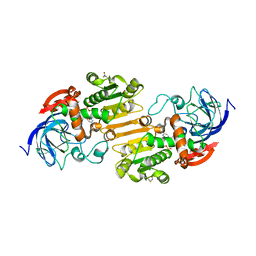 | | Crystal structure of S-nitrosylated nitrosoglutathione reductase(GSNOR)from Chlamydomonas reinhardtii, in complex with NAD+ | | 分子名称: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-(hydroxymethyl)glutathione dehydrogenase, ... | | 著者 | Fermani, S, Zaffagnini, M, Falini, G, Lemaire, S.D. | | 登録日 | 2020-11-04 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and functional insights into nitrosoglutathione reductase from Chlamydomonas reinhardtii.
Redox Biol, 38, 2020
|
|
8SY3
 
 | |
8EV1
 
 | | Dual Modulators | | 分子名称: | (3aR,4S,9bS)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3aS,4R,9bR)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, Estrogen Receptor, ... | | 著者 | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | 登録日 | 2022-10-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
6F9G
 
 | | Ligand binding domain of P. putida KT2440 polyamine chemorecpetors McpU in complex putrescine. | | 分子名称: | 1,4-DIAMINOBUTANE, ACETATE ION, GLYCEROL, ... | | 著者 | Gavira, J.A, Conejero-Muriel, M.T, Ortega, A, Martin-Mora, D, Corral-Lugo, A, Morel, B, Krell, T. | | 登録日 | 2017-12-14 | | 公開日 | 2018-03-28 | | 最終更新日 | 2018-06-13 | | 実験手法 | X-RAY DIFFRACTION (2.388 Å) | | 主引用文献 | Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida.
J. Mol. Biol., 430, 2018
|
|
4XVS
 
 | | Crystal structure of HIV-1 donor 45 d45-01dG5 coreE gp120 with antibody 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (VRC07_1995) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 Light chain, Donor 45 01dG5 coreE gp120, ... | | 著者 | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | 登録日 | 2015-01-27 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell, 161, 2015
|
|
8EV2
 
 | | Dual Modulators | | 分子名称: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | 著者 | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | 登録日 | 2022-10-19 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
7KEJ
 
 | | BDBV-289 bound to EBOV GPdMuc Makona | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab heavy chain (HC) BDBV-289, Antibody Fab light chain (LC) BDBV-289, ... | | 著者 | Murin, C.D, Ward, A.B. | | 登録日 | 2020-10-11 | | 公開日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Convergence of a common solution for broad ebolavirus neutralization by glycan cap-directed human antibodies.
Cell Rep, 35, 2021
|
|
6OPS
 
 | | HIV-1 Protease NL4-3 WT in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2019-04-25 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPW
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, V82F, I84V Mutant in complex with darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | 著者 | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | 登録日 | 2019-04-25 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
8GLO
 
 | |
6MI3
 
 | | Structure of NEMO(51-112) with N- and C-terminal coiled-coil adaptors. | | 分子名称: | NF-kB ESSENTIAL MODULATOR,NF-kappa-B essential modulator,NF-kB ESSENTIAL MODULATOR | | 著者 | Pellegrini, M, Barczewski, A.H, Mierke, D.F, Ragusa, M.J. | | 登録日 | 2018-09-19 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.783 Å) | | 主引用文献 | The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface.
Sci Rep, 9, 2019
|
|
6OV0
 
 | |
6EW7
 
 | |
6UW1
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | 分子名称: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | 著者 | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | 登録日 | 2019-11-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.205 Å) | | 主引用文献 | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6MJC
 
 | |
5J8D
 
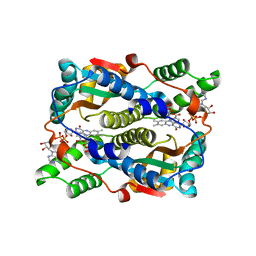 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | 分子名称: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | 著者 | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | 登録日 | 2016-04-07 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
8F1B
 
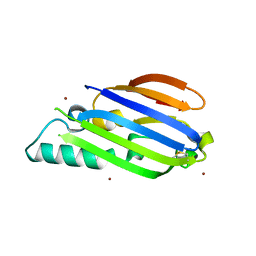 | |
8EZW
 
 | |
6MQT
 
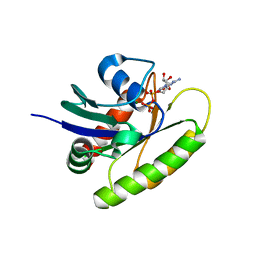 | |
8EKH
 
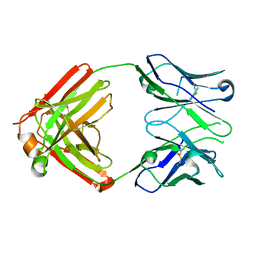 | |
6F3J
 
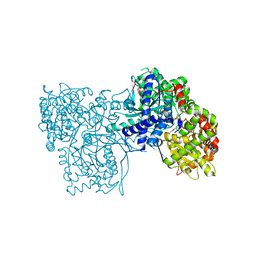 | | The crystal structure of Glycogen Phosphorylase in complex with 10a | | 分子名称: | 4-[4-[5-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-4~{H}-1,2,4-triazol-3-yl]phenyl]benzoic acid, Glycogen phosphorylase, muscle form, ... | | 著者 | Kyriakis, E, Stamati, E.C.V, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | 登録日 | 2017-11-28 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A multidisciplinary study of 3-( beta-d-glucopyranosyl)-5-substituted-1,2,4-triazole derivatives as glycogen phosphorylase inhibitors: Computation, synthesis, crystallography and kinetics reveal new potent inhibitors.
Eur J Med Chem, 147, 2018
|
|
8EOA
 
 | |
6UQE
 
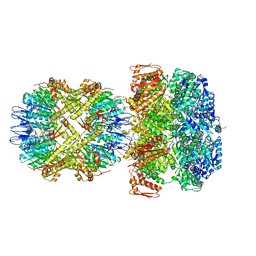 | | ClpA/ClpP Disengaged State bound to RepA-GFP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Lopez, K.L, Rizo, A.R, Southworth, D.R. | | 登録日 | 2019-10-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
