8WQY
 
 | | Fe-O nanocluster of form-XI in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WR0
 
 | | Fe-O nanocluster of form-XII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WPV
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum soaked in Fe2+ solution for 30min | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQV
 
 | | Fe-O nanocluster of form-VIII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQU
 
 | | Fe-O nanocluster of form-IX in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WQX
 
 | | Fe-O nanocluster of form-X in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, ferritin | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Wu, L.J, Wang, H.F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
9F0E
 
 | |
7P7R
 
 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
4S1X
 
 | | Crystal structure of HA2-Del-L2seM, Central Coiled-Coil from Influenza Hemagglutinin HA2 without Heptad Repeat Stutter | | Descriptor: | GLYCEROL, Truncated hemagglutinin | | Authors: | Malashkevich, V.N, Higgins, C.D, Lai, J.R, Almo, S.C. | | Deposit date: | 2015-01-15 | | Release date: | 2015-04-01 | | Last modified: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A switch from parallel to antiparallel strand orientation in a coiled-coil X-ray structure via two core hydrophobic mutations.
Biopolymers, 104, 2015
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | Descriptor: | Esterase, PHOSPHATE ION | | Authors: | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
136D
 
 | |
4RQ9
 
 | | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, GLYCEROL, ... | | Authors: | Woitowich, N.C, Halavaty, A.S, Gallagher, K.D, Nugent, A.C, Patel, H, Duong, P, Kovaleva, S.E, St.Peter, S, Ozarowski, W.B, Hernandez, C.N, Stojkovic, E.A. | | Deposit date: | 2014-10-31 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state
To be Published
|
|
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4ROB
 
 | | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, Zhang, X, Meekins, D.A, An, C, Michel, K. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8A resolution structure of SRPN2 (K198C) from Anopheles gambiae
To be Published
|
|
4TRX
 
 | |
4U1M
 
 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U1I
 
 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
1AN2
 
 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
4TW3
 
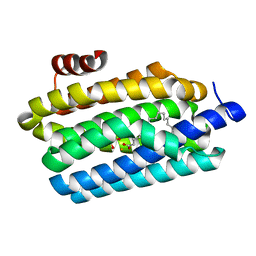 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4U2B
 
 | |
4U2E
 
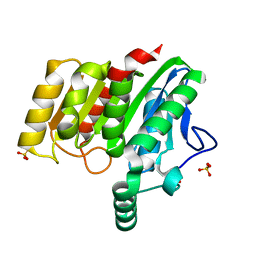 | | Crystal structure of dienelactone hydrolase S-3 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G, G211D and K234N) at 1.70 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4TW9
 
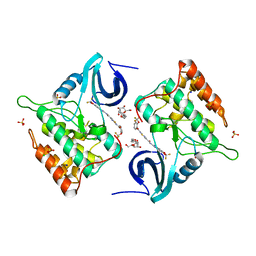 | | Difluoro-dioxolo-benzoimidazol-benzamides as potent inhibitors of CK1delta and epsilon with nanomolar inhibitory activity on cancer cell proliferation | | Descriptor: | CHLORIDE ION, Casein kinase I isoform delta, N-(2,2-difluoro-5H-[1,3]dioxolo[4,5-f]benzimidazol-6-yl)-2-{[2-(trifluoromethoxy)benzoyl]amino}-1,3-thiazole-4-carboxamide, ... | | Authors: | Richter, J, Bischof, J, Zaja, M, Kohlhof, H, Othersen, O, Vitt, D, Alscher, V, Pospiech, I, Garcia-Reyes, B, Berg, S, Leban, J, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Difluoro-dioxolo-benzoimidazol-benzamides As Potent Inhibitors of CK1 delta and epsilon with Nanomolar Inhibitory Activity on Cancer Cell Proliferation.
J.Med.Chem., 57, 2014
|
|
4U1J
 
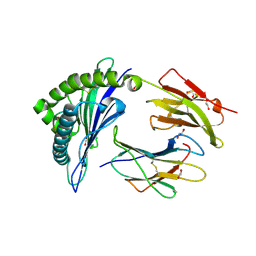 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2014-07-15 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
