2G3A
 
 | | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens | | Descriptor: | acetyltransferase | | Authors: | Cymborowski, M, Xu, X, Chruszcz, M, Zheng, H, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative acetyltransferase from Agrobacterium tumefaciens
To be Published
|
|
4KJM
 
 | | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB | | Descriptor: | ACETATE ION, CHLORIDE ION, Extracellular matrix-binding protein ebh, ... | | Authors: | Cymborowski, M, Shabalin, I.G, Joachimiak, G, Chruszcz, M, Gornicki, P, Zhang, R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB
To be Published
|
|
2IA2
 
 | | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1 | | Descriptor: | Putative Transcriptional Regulator | | Authors: | Cymborowski, M.T, Chruszcz, M, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-06 | | Release date: | 2006-10-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a putative transcriptional regulator RHA06195 from Rhodococcus sp. RHA1
To be Published
|
|
2I9C
 
 | | Crystal Structure of the Protein RPA1889 from Rhodopseudomonas palustris CGA009 | | Descriptor: | ACETATE ION, Hypothetical protein RPA1889 | | Authors: | Cymborowski, M.T, Evdokimova, E, Kagan, O, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Protein RPA1889 from Rhodopseudomonas palustris CGA009
To be Published
|
|
3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
3I4P
 
 | | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cymborowski, M, Chruszcz, M, Luo, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of AsnC family transcriptional regulator from Agrobacterium tumefaciens
To be Published
|
|
2PD0
 
 | | Protein cgd2_2020 from Cryptosporidium parvum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein | | Authors: | Cymborowski, M, Chruszcz, M, Hills, T, Lew, J, Melone, M, Zhao, Y, Artz, J, Wernimont, A, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Arrowsmith, C, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein cgd2_2020 from Cryptosporidium parvum
To be Published
|
|
2FEF
 
 | | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, hypothetical protein PA2201 | | Authors: | Cymborowski, M.T, Chruszcz, M, Wang, S, Shumilin, I, Kudritska, M, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Protein PA2201 from Pseudomonas aeruginosa
To be Published
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2Q24
 
 | | Crystal structure of TetR transcriptional regulator SCO0520 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative tetR family transcriptional regulator | | Authors: | Cymborowski, M, Chruszcz, M, Koclega, K.D, Filippova, E.V, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-25 | | Release date: | 2007-07-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
J.Struct.Funct.Genom., 12, 2011
|
|
4WCZ
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
4TNN
 
 | | Crystal structure of Escherichia coli protein YodA in complex with Ni - artifact of purification. | | Descriptor: | Metal-binding lipocalin, NICKEL (II) ION, SULFATE ION | | Authors: | Gasiorowska, O.A, Cymborowski, M.T, Handing, K.B, Shabalin, I.G, Zasadzinska, E, Niedzialkowska, E, Porebski, P.J, Minor, W. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
2G7U
 
 | | 2.3 A structure of putative catechol degradative operon regulator from Rhodococcus sp. RHA1 | | Descriptor: | transcriptional regulator | | Authors: | Zheng, H, Skarina, T, Chruszcz, M, Cymborowski, M, Grabowski, M, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 A structure of putative catechol degradative operon regulator from
Rhodococcus sp. RHA1
To be Published
|
|
2G8Y
 
 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
2G3B
 
 | | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | GLYCEROL, putative TetR-family transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Cymborowski, M, Kagan, O, Wang, S, Koclega, K.D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative TetR-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
6S0R
 
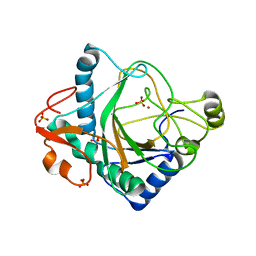 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | Descriptor: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
5IIU
 
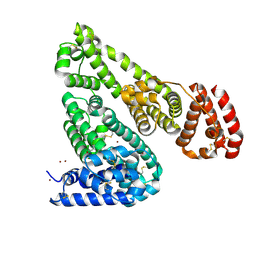 | | Crystal structure of Equine Serum Albumin in the presence of 10 mM zinc at pH 6.9 | | Descriptor: | SULFATE ION, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Shabalin, I.G, Cooper, D.R, Cymborowski, M.T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
4WED
 
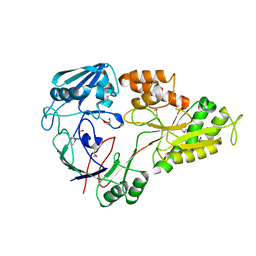 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | Descriptor: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | Authors: | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
5BP9
 
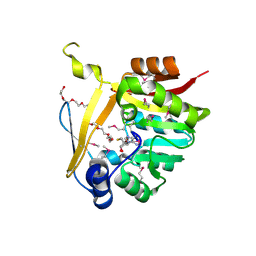 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
5TX7
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Czub, M.P, Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Kutner, J, Cymborowski, M.T, Hennig, P.M, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-11-15 | | Release date: | 2016-12-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase from Desulfovibrio vulgaris
to be published
|
|
4ZNZ
 
 | | Crystal structure of Escherichia coli carbonic anhydrase (YadF) in complex with Zn - artifact of purification | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Gasiorowska, O.A, Niedzialkowska, E, Porebski, P.J, Handing, K.B, Shabalin, I.G, Cymborowski, M.T, Minor, W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4XOX
 
 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
1T9K
 
 | | X-ray crystal structure of aIF-2B alpha subunit-related translation initiation factor [Thermotoga maritima] | | Descriptor: | CHLORIDE ION, Probable methylthioribose-1-phosphate isomerase, SULFATE ION | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-17 | | Release date: | 2004-06-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of aIF-2B translation initiation factor from Thermotoga maritima
To be Published
|
|
5IJ5
 
 | | Crystal structure of Equine Serum Albumin in the presence of 50 mM zinc at pH 4.5 | | Descriptor: | GLYCEROL, Serum albumin, ZINC ION | | Authors: | Handing, K.B, Majorek, K.A, Shabalin, I.G, Cymborowski, M.T, Zheng, H, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Circulatory zinc transport is controlled by distinct interdomain sites on mammalian albumins.
Chem Sci, 7, 2016
|
|
