5WOO
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with Myristic acid (C14:0) to 1.78 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6DJ6
 
 | | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with cis-oleic acid to 1.9 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B2 protein (SPR1019), GLYCEROL, OLEIC ACID, ... | | Authors: | Cuypers, M.G, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-05-24 | | Release date: | 2019-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein (SPR1019) loaded with cis-oleic acid to 1.9 Angstrom resolution
To Be Published
|
|
6DKE
 
 | |
6WS3
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomers TG and AGCA (from cleaved GTGAGCAGTG) | | Descriptor: | DNA (5'-D(P*TP*G)-3'), GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7LP8
 
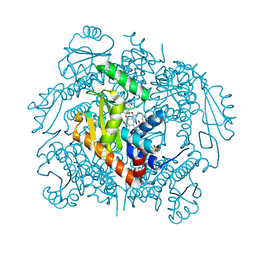 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
7LP7
 
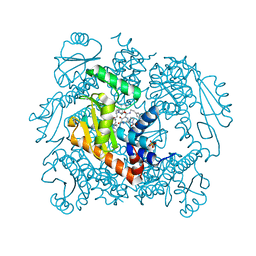 | | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476 | | Descriptor: | (phenylmethyl) (2~{S})-2-[5-oxidanyl-6-oxidanylidene-4-(2-pyridin-4-ylethylcarbamoyl)-1~{H}-pyrimidin-2-yl]pyrrolidine-1-carboxylate, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000983476
To Be Published
|
|
6TGT
 
 | |
6TB5
 
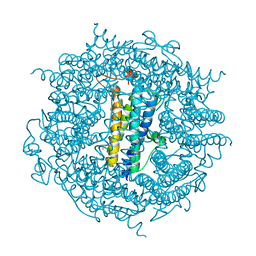 | | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h). | | Descriptor: | CALCIUM ION, DNA protection during starvation protein 2, FE (III) ION | | Authors: | Cuypers, M.G, McSweeney, S, Romao, C.V, Mitchell, E.P. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the DPS2 from DEINOCOCCUS RADIODURANS to 1.83A resolution (sequentially soaked in CaCl2 [5mM] for 20 min, then in Ammonium iron(II) sulfate [10mM] for 2h).
To Be Published
|
|
5AI3
 
 | |
5AI2
 
 | |
6WIJ
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant I38T in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6WJ4
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type in complex with SJ000986448 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7LW6
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T mutant in complex with Raltegravir | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, White, S.W, Rankovik, Z. | | Deposit date: | 2021-02-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6ALW
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 12-Methyl Myristic Acid (C15:0) to 1.63 Angstrom resolution | | Descriptor: | (12R)-12-methyltetradecanoic acid, (12S)-12-methyltetradecanoic acid, EDD domain protein, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6B9I
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 14-Methylhexadecanoic Acid (Anteiso C17:0) to 1.93 Angstrom resolution | | Descriptor: | (14S)-14-methylhexadecanoic acid, Fatty acid Kinase (Fak) B1, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
7UQ1
 
 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and a single Mg ion at the dinuclear binding site | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fatty Acid Kinase A, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, Seetharaman, J, Rock, C.O, White, S.W. | | Deposit date: | 2022-04-18 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Domain architecture and catalysis of the Staphylococcus aureus fatty acid kinase.
J.Biol.Chem., 298, 2022
|
|
7UUH
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001034732-2 (cis-form) | | Descriptor: | (1P,18Z)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2(28),5,11(27),12,14,18,22,24-nonaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6CNG
 
 | | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B3 protein loaded with linoleic acid to 1.47 Angstrom resolution | | Descriptor: | ACETATE ION, Fatty Acid Kinase (Fak) B3 protein, GLYCEROL, ... | | Authors: | Cuypers, M.G, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-03-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The X-ray crystal structure of the Streptococcus pneumoniae Fatty Acid Kinase (Fak) B3 protein loaded with linoleic acid to 1.47 Angstrom resolution
To Be Published
|
|
7UMR
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001034732-1 (trans-form) | | Descriptor: | (1P,18Z)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2(28),5,11(27),12,14,18,22,24-nonaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5JLT
 
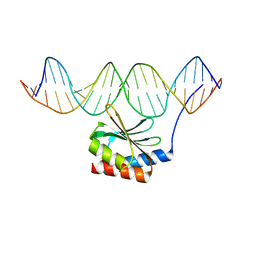 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
7RKP
 
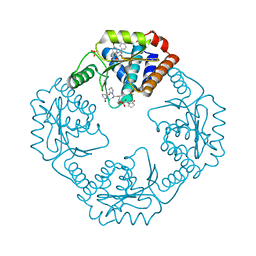 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with cyclic compound SJ001034733 | | Descriptor: | (5R)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2,6(28),11(27),12,14,22,24-octaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7K77
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ001008025
To Be Published
|
|
7KAF
 
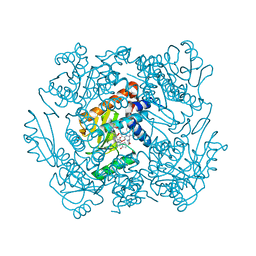 | | The crystal structure of the 2009/H1N1/California PA endonuclease I38T (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-09-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
7KBC
 
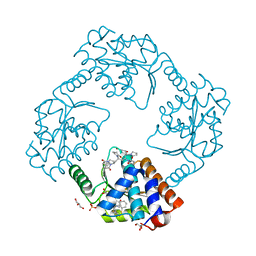 | | The crystal structure of the 2009/H1N1/California PA endonuclease mutant E119D (construct with truncated loop 51-72) in complex with baloxavir acid | | Descriptor: | Baloxavir acid, GLYCEROL, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Kumar, G, Slavish, J, White, S.W. | | Deposit date: | 2020-10-02 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
