1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
3UQ3
 
 | | TPR2AB-domain:pHSP90-complex of yeast Sti1 | | Descriptor: | Heat shock protein, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
3UDG
 
 | | Structure of Deinococcus radiodurans SSB bound to ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein, THYMIDINE-5'-PHOSPHATE | | Authors: | George, N.P, Ngo, K.V, Chitteni-Patu, S, Norais, C.A, Battista, J.R, Cox, M.M, Keck, J.L. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes.
J.Biol.Chem., 287, 2012
|
|
5JRJ
 
 | | Crystal Structure of Herbaspirillum seropedicae RecA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Leite, W.C, Galvao, C.W, Saab, S.C, Iulek, J, Etto, R.M, Steffens, M.B.R, Chitteni-Pattu, S, Stanage, T, Keck, J.L, Cox, M.M. | | Deposit date: | 2016-05-06 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of H. seropedicae RecA Protein - Insights into the Polymerization of RecA Protein as Nucleoprotein Filament.
PLoS ONE, 11, 2016
|
|
4H5B
 
 | | Crystal Structure of DR_1245 from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, DR_1245 protein, GLYCEROL, ... | | Authors: | Norais, C, Servant, P, Bouthier-de-la-Tour, C, Coureux, P.D, Ithurbide, S, Vannier, F, Guerin, P, Dulberger, C.L, Satyshur, K.A, Keck, J.L, Armengaud, J, Cox, M.M, Sommer, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Deinococcus radiodurans DR1245 Protein, a DdrB Partner Homologous to YbjN Proteins and Reminiscent of Type III Secretion System Chaperones.
Plos One, 8, 2013
|
|
7R7J
 
 | | Crystal structure of RadD with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | Authors: | Osorio Garcia, M.A, Satyshur, K.A, Keck, J.L, Cox, M.M. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the Escherichia coli RadD DNA repair protein bound to ADP reveals a novel zinc ribbon domain.
Plos One, 17, 2022
|
|
3NCT
 
 | | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of reca | | Descriptor: | Protein psiB | | Authors: | Petrova, V, Satyshur, K.A, George, N.P, McCaslin, D, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of RecA.
J.Biol.Chem., 285, 2010
|
|
3PLW
 
 | | Ref protein from P1 bacteriophage | | Descriptor: | Recombination enhancement function protein, SULFATE ION, ZINC ION | | Authors: | Keck, J.L, Lu, D, Cox, M.M. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Creating Directed Double-strand Breaks with the Ref Protein: A NOVEL RecA-DEPENDENT NUCLEASE FROM BACTERIOPHAGE P1.
J.Biol.Chem., 286, 2011
|
|
1SE8
 
 | | Structure of single-stranded DNA-binding protein (SSB) from D. radiodurans | | Descriptor: | Single-strand binding protein | | Authors: | Bernstein, D.A, Eggington, J.M, Killoran, M.P, Misic, A.M, Cox, M.M, Keck, J.L. | | Deposit date: | 2004-02-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3PVS
 
 | | Structure and biochemical activities of Escherichia coli MgsA | | Descriptor: | PHOSPHATE ION, Replication-associated recombination protein A | | Authors: | Page, A.N, George, N.P, Marceau, A.H, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-12-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Biochemical Activities of Escherichia coli MgsA.
J.Biol.Chem., 286, 2011
|
|
2LLV
 
 | | Solution structure of the yeast Sti1 DP1 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
2LLW
 
 | | Solution structure of the yeast Sti1 DP2 domain | | Descriptor: | Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H, Buchner, J. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
3UPV
 
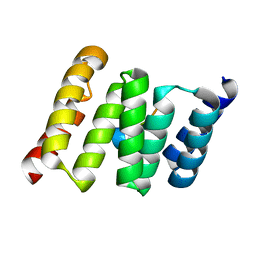 | | TPR2B-domain:pHsp70-complex of yeast Sti1 | | Descriptor: | Heat shock protein SSA4, Heat shock protein STI1 | | Authors: | Schmid, A.B, Lagleder, S, Graewert, M.A, Roehl, A, Hagn, F, Wandinger, S.K, Cox, M.B, Demmer, O, Richter, K, Groll, M, Kessler, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop.
Embo J., 31, 2012
|
|
1FLO
 
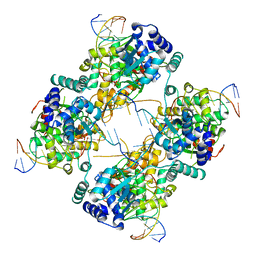 | | FLP Recombinase-Holliday Junction Complex I | | Descriptor: | FLP RECOMBINASE, PHOSPHONIC ACID, SYMMETRIZED FRT DNA SITES | | Authors: | Chen, Y, Narendra, U, Iype, L.E, Cox, M.M, Rice, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping.
Mol.Cell, 6, 2000
|
|
2B8H
 
 | | A/NWS/whale/Maine/1/84 (H1N9) reassortant influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Smith, B.J, Platis, D, Cox, M.M.J, Huyton, T, Joosten, R.P, McKimm-Breschkin, J.L, Zhang, J.-G, Luo, C.S, Lou, M.-Z, Garrett, T.P.J, Labrou, N.E. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4U0P
 
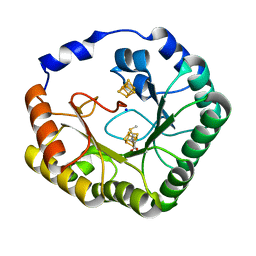 | | The Crystal Structure of Lipoyl Synthase in Complex with S-Adenosyl Homocysteine | | Descriptor: | IRON/SULFUR CLUSTER, Lipoyl synthase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Sandy, J, Dinis, P.C, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
4U0O
 
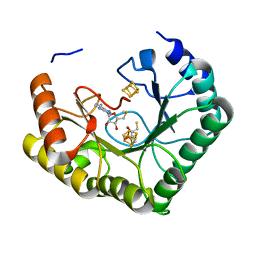 | | Crystal structure of Thermosynechococcus elongatus Lipoyl Synthase 2 complexed with MTA and DTT | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Harmer, J.E, Hiscox, M.J, Dinis, P.C, Sandy, J, Roach, P.L. | | Deposit date: | 2014-07-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions.
Biochem.J., 464, 2014
|
|
1P2A
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a trisubstituted naphthostyril inhibitor | | Descriptor: | 5-[(2-AMINOETHYL)AMINO]-6-FLUORO-3-(1H-PYRROL-2-YL)BENZO[CD]INDOL-2(1H)-ONE, Cell division protein kinase 2 | | Authors: | Liu, J.-J, Dermatakis, A, Lukacs, C.M, Konzelmann, F, Chen, Y, Kammlott, U, Depinto, W, Yang, H, Yin, X, Chen, Y, Schutt, A, Simcox, M.E, Luk, K.-C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3,5,6-Trisubstituted Naphthostyrils as CDK2 Inhibitors
BIOORG.MED.CHEM., 13, 2003
|
|
1R78
 
 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
