8EB0
 
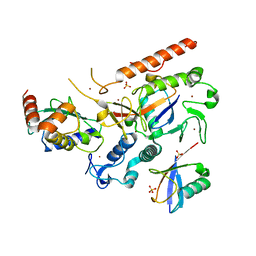 | | RNF216/E2-Ub/Ub transthiolation complex | | Descriptor: | E3 ubiquitin-protein ligase RNF216, SULFATE ION, Ubiquitin, ... | | Authors: | Cotton, T.R, Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
7M4O
 
 | |
7M4M
 
 | |
7M4N
 
 | |
5NTU
 
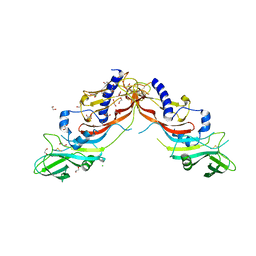 | | Crystal Structure of human Pro-myostatin Precursor at 2.6 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Growth/differentiation factor 8 | | Authors: | Cotton, T.R, Fischer, G, Hyvonen, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human myostatin precursor and determinants of growth factor latency.
EMBO J., 37, 2018
|
|
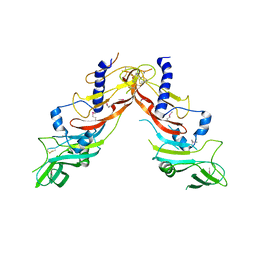
5NXS
 
 | |
8EAZ
 
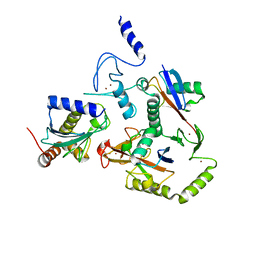 | | HOIL-1/E2-Ub/Ub transthiolation complex | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Wang, X.S, Cotton, T.R, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
