1AW5
 
 | |
1OEX
 
 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1GN6
 
 | | G152A mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Saward, S, Erskine, P.T, Badasso, M.O, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-Ray Structure Analysis of an Engineered Fe-Superoxide Dismutase Gly-Ala Mutant with Significantly Reduced Stability to Denaturant
FEBS Lett., 387, 1996
|
|
1H4J
 
 | | Methylobacterium extorquens methanol dehydrogenase D303E mutant | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 1, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Mohammed, F, Gill, R, Thompson, D, Cooper, J.B, Wood, S.P, Afolabi, P.R, Anthony, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Directed Mutagenesis and X-Ray Crystallography of the Pqq-Containing Quinoprotein Methanol Dehydrogenase and its Electron Acceptor, Cytochrome C(L)(,)
Biochemistry, 40, 2001
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1B4E
 
 | | X-ray structure of 5-aminolevulinic acid dehydratase complexed with the inhibitor levulinic acid | | Descriptor: | GLYCEROL, LAEVULINIC ACID, PROTEIN (5-AMINOLEVULINIC ACID DEHYDRATASE), ... | | Authors: | Erskine, P.T, Cooper, J.B, Lewis, G, Spencer, P, Wood, S.P, Shoolingin-Jordan, P.M. | | Deposit date: | 1998-12-19 | | Release date: | 1999-12-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 5-aminolevulinic acid dehydratase from Escherichia coli complexed with the inhibitor levulinic acid at 2.0 A resolution.
Biochemistry, 38, 1999
|
|
1W1Z
 
 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
2VS2
 
 | | Neutron diffraction structure of endothiapepsin in complex with a gem- diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
3URI
 
 | | Endothiapepsin-DB5 complex. | | Descriptor: | DB5 peptide, Endothiapepsin | | Authors: | Bailey, D, Sanz-Aparicio, J, Albert, A, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3URL
 
 | | Endothiapepsin-DB6 complex. | | Descriptor: | DB6 peptide, Endothiapepsin, SULFATE ION | | Authors: | Bailey, D, Sanz-Aparicio, J, Albert, A, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1EB3
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1RRB
 
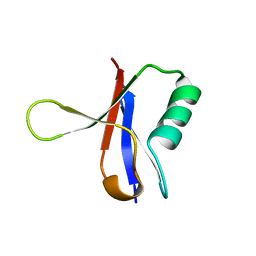 | | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | | Descriptor: | RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Terada, T, Ito, Y, Shirouzu, M, Tateno, M, Hashimoto, K, Kigawa, T, Ebisuzaki, T, Takio, K, Shibata, T, Yokoyama, S, Smith, B.O, Laue, E.D, Cooper, J.A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance and molecular dynamics studies on the interactions of the Ras-binding domain of Raf-1 with wild-type and mutant Ras proteins.
J.Mol.Biol., 286, 1999
|
|
3NFT
 
 | | Near-atomic resolution analysis of BipD- A component of the type-III secretion system of Burkholderia pseudomallei | | Descriptor: | Translocator protein bipD | | Authors: | Pal, M, Erskine, P.T, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Near-atomic resolution analysis of BipD, a component of the type III secretion system of Burkholderia pseudomallei.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3URJ
 
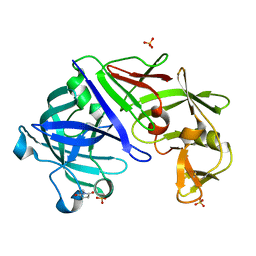 | | Type IV native endothiapepsin | | Descriptor: | Endothiapepsin, SULFATE ION | | Authors: | Bailey, D, Cooper, J.B. | | Deposit date: | 2011-11-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UTL
 
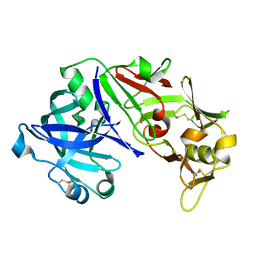 | | Human pepsin 3b | | Descriptor: | Pepsin A | | Authors: | Coker, A, Cooper, J.B. | | Deposit date: | 2011-11-25 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An analysis of subdomain orientation, conformational change and disorder in relation to crystal packing of aspartic proteinases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2WYX
 
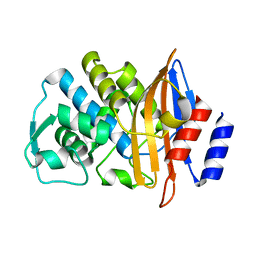 | | Neutron structure of a class A Beta-lactamase Toho-1 E166A R274N R276N triple mutant | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Blakeley, M.P, Cooper, J, Chen, Y, Afonine, P, Coates, L. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Neutron Diffraction Studies of a Class a Beta-Lactamase Toho-1 E166A R274N R276N Triple Mutant
J.Mol.Biol., 396, 2010
|
|
1E51
 
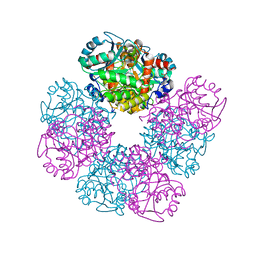 | | Crystal structure of native human erythrocyte 5-aminolaevulinic acid dehydratase | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, SULFATE ION, ... | | Authors: | Mills-Davies, N.L, Thompson, D, Cooper, J.B, Shoolingin-Jordan, P.M. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Human Ala-Dehydratase
To be Published
|
|
1GKT
 
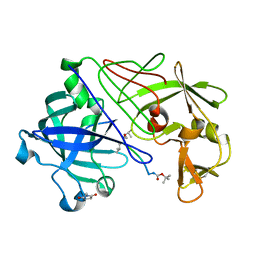 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
2BJI
 
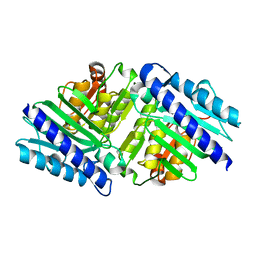 | | High Resolution Structure of myo-Inositol Monophosphatase, The Target of Lithium Therapy | | Descriptor: | INOSITOL-1(OR 4)-MONOPHOSPHATASE, MAGNESIUM ION | | Authors: | Gill, R, Mohammed, F, Badyal, R, Coates, L, Erskine, P, Thompson, D, Cooper, J, Gore, M, Wood, S. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
2XR0
 
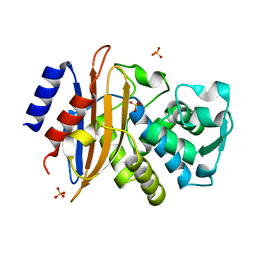 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
2XQZ
 
 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
1W6S
 
 | | The high resolution structure of methanol dehydrogenase from methylobacterium extorquens | | Descriptor: | CALCIUM ION, GLYCEROL, METHANOL DEHYDROGENASE SUBUNIT 1, ... | | Authors: | Williams, P.A, Coates, L, Mohammed, F, Gill, R, Erskine, P.T, Wood, S.P, Anthony, C, Cooper, J.B. | | Deposit date: | 2004-08-23 | | Release date: | 2004-12-21 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Atomic Resolution Structure of Methanol Dehydrogenase from Methylobacterium Extorquens
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C1H
 
 | | The X-ray Structure of Chlorobium vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor | | Descriptor: | 4,7-DIOXOSEBACIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S, Wood, S.P, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2005-09-14 | | Release date: | 2005-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Chlorobium Vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5NFE
 
 | | Neutron structure of human transthyretin (TTR) T119M mutant at room temperature to 1.85A resolution | | Descriptor: | Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Ostermann, A, Cooper, J.B, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2017-03-14 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.853 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
