4U2B
 
 | |
4U2E
 
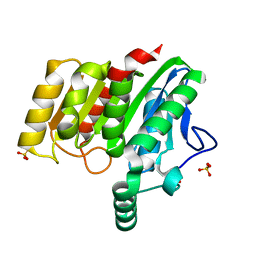 | | Crystal structure of dienelactone hydrolase S-3 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G, G211D and K234N) at 1.70 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2F
 
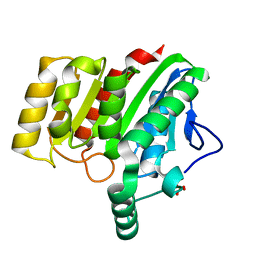 | | Crystal structure of dienelactone hydrolase B-1 variant (Q35H, F38L, Y64H, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2C
 
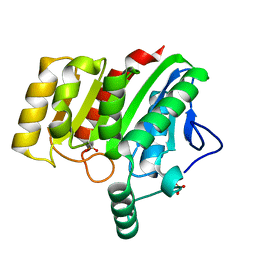 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2D
 
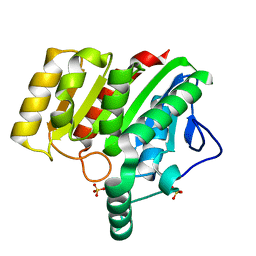 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2G
 
 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4P93
 
 | | Structure of Dienelactone Hydrolase at 1.85 A resolution crystallised in the C2 space group | | Descriptor: | Carboxymethylenebutenolidase | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4P92
 
 | | Crystal structure of dienelactone hydrolase C123S mutant at 1.65 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Carr, P.D, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization of dienelactone hydrolase in two space groups: structural changes caused by crystal packing.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3KM5
 
 | | Crystal Structure Analysis of the K2 Cleaved Adhesin Domain of Lys-gingipain (Kgp) | | Descriptor: | CALCIUM ION, GLYCEROL, Lysine specific cysteine protease, ... | | Authors: | Li, N, Collyer, C.A, Hunter, N. | | Deposit date: | 2009-11-09 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis
Mol.Microbiol., 76, 2010
|
|
2EPL
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | GLYCEROL, N-acetyl-beta-D-glucosaminidase, SULFATE ION | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2EPM
 
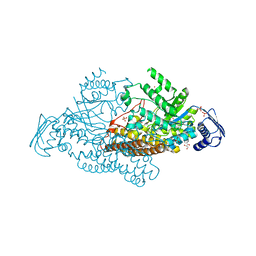 | | N-acetyl-B-D-glucoasminidase (GCNA) from Stretococcus gordonii | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-acetyl-beta-D-glucosaminidase, ... | | Authors: | Langley, D.B, Harty, D.W.S, Guss, J.M, Collyer, C.A. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2DFY
 
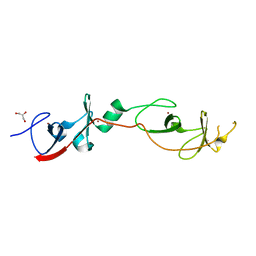 | | Crystal structure of a cyclized protein fusion of LMO4 LIM domains 1 and 2 with the LIM interacting domain of LDB1 | | Descriptor: | GLYCEROL, ZINC ION, fusion protein of LIM domain transcription factor LMO4 and LIM domain-binding protein 1 | | Authors: | Jeffries, C.M.J, Graham, S.C, Collyer, C.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of a binary protein complex by intein-mediated cyclization
Protein Sci., 15, 2006
|
|
1DUV
 
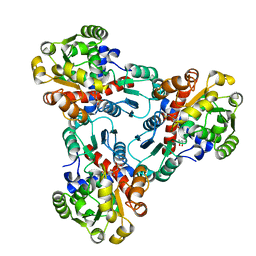 | | CRYSTAL STRUCTURE OF E. COLI ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH NDELTA-L-ORNITHINE-DIAMINOPHOSPHINYL-N-SULPHONIC ACID (PSORN) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDELTA-(N'-SULPHODIAMINOPHOSPHINYL)-L-ORNITHINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Langley, D.B, Templeton, M.D, Fields, B.A, Mitchell, R.E, Collyer, C.A. | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N'-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism.
J.Biol.Chem., 275, 2000
|
|
1DL8
 
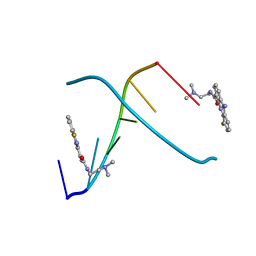 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
