8BYJ
 
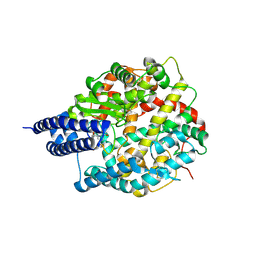 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
5C6F
 
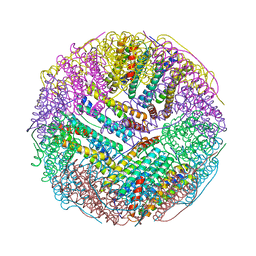 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
6SPN
 
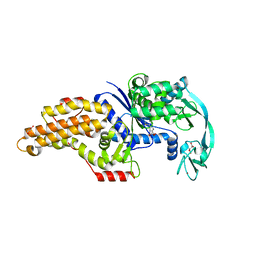 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6JCE
 
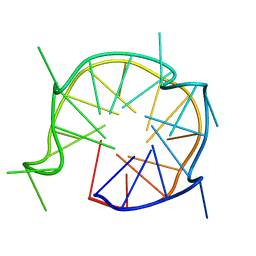 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
5MLQ
 
 | | Structure of CDPS from Nocardia brasiliensis | | Descriptor: | CDPS, CITRIC ACID | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
6EZ3
 
 | |
6FQ2
 
 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5L4O
 
 | |
6GZ6
 
 | | Structure of a left-handed G-quadruplex | | Descriptor: | DNA (27-MER), POTASSIUM ION | | Authors: | Bakalar, B, Heddi, B, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-07-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5OCD
 
 | |
5MLP
 
 | | Structure of CDPS from Rickettsiella grylli | | Descriptor: | Uncharacterized protein | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
8PHV
 
 | | Structure of Human selenomethionylated Cdc123 bound to domain 3 of eIF2 gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123 homolog, Eukaryotic translation initiation factor 2 subunit 3 | | Authors: | Schmitt, E, Mechulam, Y, Cardenal Peralta, C, Fagart, J, Seufert, W. | | Deposit date: | 2023-06-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of human Cdc123 to eIF2 gamma.
J.Struct.Biol., 215, 2023
|
|
8PHD
 
 | | Structure of Human Cdc123 bound to domain 3 of eIF2 gamma and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123 homolog, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Schmitt, E, Mechulam, Y, Cardenal Peralta, C, Fagart, J, Seufert, W. | | Deposit date: | 2023-06-19 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Binding of human Cdc123 to eIF2 gamma.
J.Struct.Biol., 215, 2023
|
|
1KK0
 
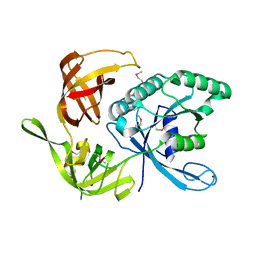 | |
1KK3
 
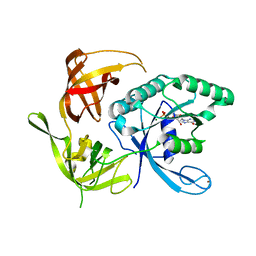 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KK2
 
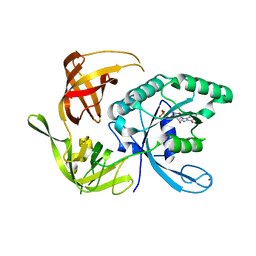 | | Structure of the large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi-G235D mutant complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KJZ
 
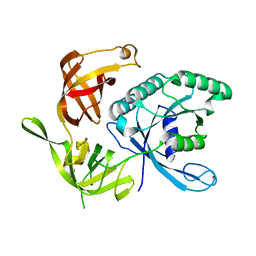 | |
1KK1
 
 | | Structure of the large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi-G235D mutant complexed with GDPNP-Mg2+ | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
4Q24
 
 | | Crystal structure of Cyclo(L-leucyl-L-phenylalanyl) synthase | | Descriptor: | Cyclo(L-leucyl-L-phenylalanyl) synthase, PHENYLMETHYL N-[(2S)-4-CHLORO-3-OXO-1-PHENYL-BUTAN-2-YL]CARBAMATE | | Authors: | Moutiez, M, Schmitt, E, Seguin, J, Thai, R, Favry, E, Mechulam, Y, Gondry, M. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases.
Nat Commun, 5, 2014
|
|
6SPR
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SPP
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
4ZGQ
 
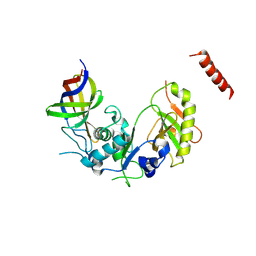 | | Structure of Cdc123 bound to eIF2-gammaDIII domain | | Descriptor: | Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
4ZGN
 
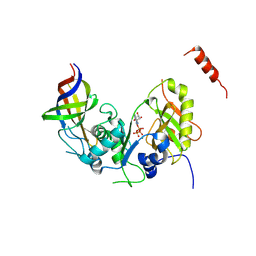 | | Structure Cdc123 complexed with the C-terminal domain of eIF2gamma | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
6SWE
 
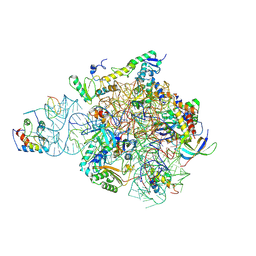 | | IC2 head of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SWC
 
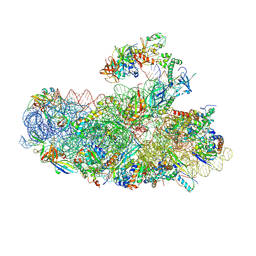 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
