4HI7
 
 | | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione | | Descriptor: | GI20122, GLUTATHIONE | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-11 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione
To be Published
|
|
4NX1
 
 | | Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate | | Descriptor: | C4-dicarboxylate transport system substrate-binding protein, alpha-D-talopyranuronic acid | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4G8T
 
 | | Crystal structure of a glucarate dehydratase related protein, from actinobacillus succinogenes, target EFI-502312, with sodium and sulfate bound, ordered loop | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glucarate dehydratase related protein, from actinobacillus succinogenes, target efi-502312, with sodium and sulfate bound, ordered loop
TO BE PUBLISHED
|
|
4GIS
 
 | | crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, glycerol and dicarboxylates bound (mixed loops, space group I4122) | | Descriptor: | CHLORIDE ION, Enolase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, glycerol and dicarboxylates bound (mixed loops, space group I4122)
To be Published
|
|
4H83
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing enzyme (EFI target:502127) | | Descriptor: | BICARBONATE ION, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing enzyme
To be Published
|
|
4R2F
 
 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | Descriptor: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
4QSE
 
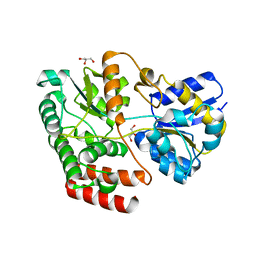 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum c58, target efi-510558, with bound glycerol | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, K.L, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of maltoside transporter ATU4361 from
Agrobacterium Fabrum, target EFI-510558
To be Published
|
|
4R6Y
 
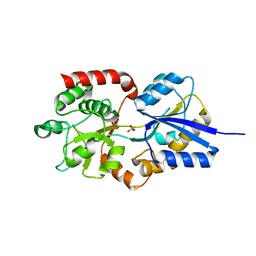 | | Crystal structure of solute-binding protein stm0429 from salmonella enterica subsp. enterica serovar typhimurium str. lt2, target efi-510776, a closed conformation, in complex with glycerol and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Putative 2-aminoethylphosphonate-binding periplasmic protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of Transporter STM0429 from Salmonella Enterica
To be Published
|
|
4QSC
 
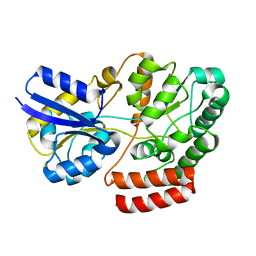 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum C58, target efi-510558, with bound maltose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Maltoside Transporter Atu4361 from Agrobacterium Fabrum, Target Efi-510558
To be Published
|
|
4HOJ
 
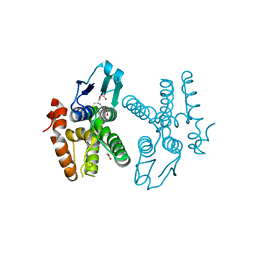 | | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione | | Descriptor: | ACETATE ION, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of glutathione transferase homolog from Neisseria Gonorrhoeae, target EFI-501841, with bound glutathione
To be Published
|
|
4NKR
 
 | | The Crystal structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Choe, J, Kim, D, Choi, S, Kim, H. | | Deposit date: | 2013-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insight into the Role of E.coli MobB in Molybdenum Cofactor Biosynthesis based on the high resolution crystal structure
TO BE PUBLISHED
|
|
4GI5
 
 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
2WTF
 
 | | DNA polymerase eta in complex with the cis-diammineplatinum (II) 1,3- GTG intrastrand cross-link | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*TP*GP*GP*TP*GP*AP*GP*CP)-3', 5'-D(*TP*CP*TP*TP*CP*TP*GP*TP*GP*CP *TP*CP*AP*CP*CP*AP*CP)-3', ... | | Authors: | Reissner, T, Schneider, S, Ziv, O, Schorr, S, Livneh, Z, Carell, T. | | Deposit date: | 2009-09-16 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Cisplatin-(1,3-Gtg) Cross-Link within DNA Polymerase Eta.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2WVO
 
 | | Structure of the HET-S N-terminal domain | | Descriptor: | CHLORIDE ION, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
2X1N
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
7KX7
 
 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*AP*GP*(OMC))-3') | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
7KX9
 
 | | Cryo-EM structure of Ephydatia fluviatilis PiwiA-piRNA-target complex | | Descriptor: | MAGNESIUM ION, Piwi-A, RNA (5'-R(P*UP*CP*UP*CP*UP*UP*GP*AP*GP*UP*UP*GP*GP*AP*CP*AP*AP*AP*UP*GP*GP*CP*AP*(OMG))-3'), ... | | Authors: | Anzelon, T.A, Chowdhury, S, Lander, G.C, MacRae, I.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for piRNA targeting.
Nature, 597, 2021
|
|
2XGQ
 
 | | Structure of yeast DNA polymerase eta in complex with C8-N-acetyl-2- aminoanthracene containing DNA | | Descriptor: | 5'-D(*CP*8AG*CP*TP*CP*AP*TP*CP*CP*AP*C)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*AP*G)-3', CALCIUM ION, ... | | Authors: | Schneider, S, Schorr, S, Carell, T. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Replication Blocking and Bypass of Y-Family Polymerase Eta by Bulky Acetylaminofluorene DNA Adducts.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N17
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, With bound beta-D-galacturonate | | Descriptor: | CALCIUM ION, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4GP2
 
 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP and Magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
4GP1
 
 | | Crystal structure of ISOPRENOID SYNTHASE A3MSH1 (TARGET EFI-501992) from pyrobaculum calidifontis complexed with DMAPP | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, Polyprenyl synthetase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase from Pyrobaculum Calidifontis
To be Published
|
|
4GIC
 
 | | Crystal Structure Of a Putative Histidinol dehydrogenase (Target PSI-014034) from Methylococcus capsulatus | | Descriptor: | Histidinol dehydrogenase, SULFATE ION | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a putative Histidinol dehydrogenase from Methylococcus capsulatus
to be published
|
|
4HWU
 
 | | Crystal structure of the Ig-C2 type 1 domain from mouse Fibroblast growth factor receptor 2 (FGFR2) [NYSGRC-005912] | | Descriptor: | Fibroblast growth factor receptor 2 | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of the Ig-C2 type 1 domain from mouse FGFR2 [NYSGRC-005912]
to be published
|
|
4ECJ
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
