1B8M
 
 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
3UM6
 
 | | Double mutant (A16V+S108T) Plasmodium falciparum DHFR-TS (T9/94) complexed with cycloguanil, NADPH and dUMP | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
2LOR
 
 | | Backbone structure of human membrane protein TMEM141 | | Descriptor: | Transmembrane protein 141 | | Authors: | Bayrhuber, M, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LON
 
 | |
2LOO
 
 | | Backbone structure of human membrane protein TMEM14A from NOE data | | Descriptor: | Transmembrane protein 14A | | Authors: | Eichmann, C, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOP
 
 | | Backbone structure of human membrane protein TMEM14A | | Descriptor: | Transmembrane protein 14A | | Authors: | Eichmann, C, Klammt, C, Maslennikov, I, Kwiatkowski, W, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
4XFR
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group P6522 | | Descriptor: | CITRIC ACID, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-28 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XFM
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, with bound D-threonate, domain swapped dimer | | Descriptor: | THREONATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-27 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XGJ
 
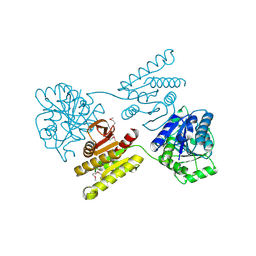 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, APO structure, domain swapped dimer | | Descriptor: | Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3TWB
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and gluconic acid | | Descriptor: | CHLORIDE ION, D-gluconic acid, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
4Y1H
 
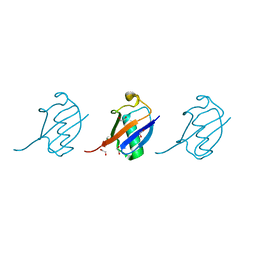 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4XFE
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, SULFATE ION, TRAP dicarboxylate transporter subunit DctP, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate
To be published
|
|
3TWA
 
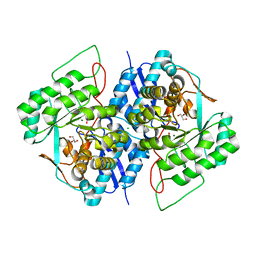 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
1LSU
 
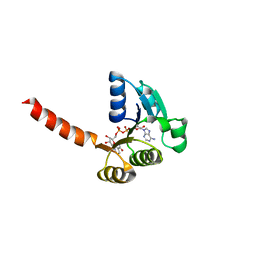 | | KTN Bsu222 Crystal Structure in Complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Conserved hypothetical protein yuaA | | Authors: | Roosild, T.P, Miller, S, Booth, I.R, Choe, S. | | Deposit date: | 2002-05-18 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch.
Cell(Cambridge,Mass.), 109, 2002
|
|
2E11
 
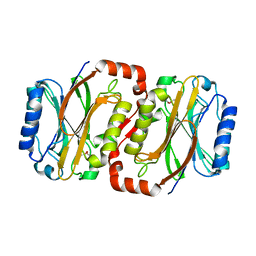 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
4XG0
 
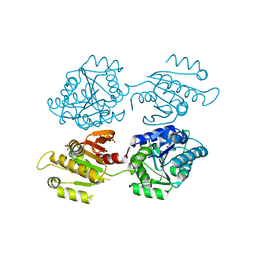 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group C2221 | | Descriptor: | CHLORIDE ION, CITRIC ACID, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3TS7
 
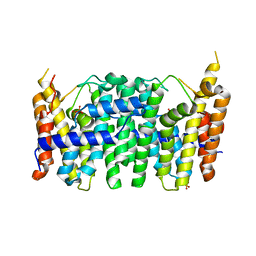 | | CRYSTAL STRUCTURE OF FARNESYL DIPHOSPHATE SYNTHASE (TARGET EFI-501951) FROM Methylococcus capsulatus | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TW9
 
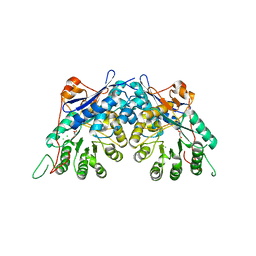 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative dehydratase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
2L2X
 
 | | Thiostrepton, oxidized at CA-CB bond of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2WVQ
 
 | | Structure of the HET-s N-terminal domain. Mutant D23A, P33H | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of prion inhibition by HET-S.
Mol. Cell, 38, 2010
|
|
4XYZ
 
 | | Crystal structure of K33 linked di-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
4WGH
 
 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
2AQA
 
 | |
4F62
 
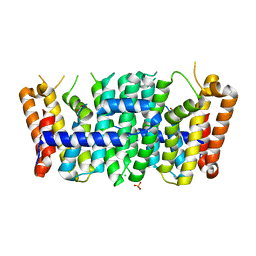 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MMR
 
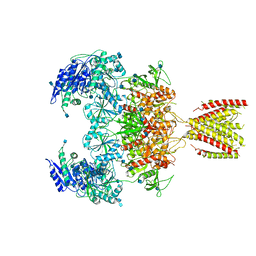 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar zinc chloride, 3 millimolar EDTA, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.13 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
