7ZLV
 
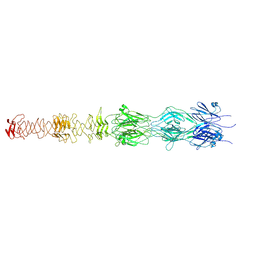 | | Tail tip of siphophage T5 : central fibre protein pb4 | | Descriptor: | Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-15 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
8B14
 
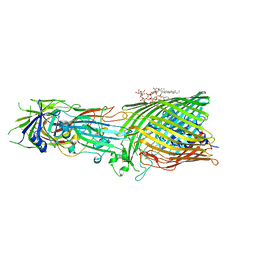 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | Authors: | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | Deposit date: | 2022-09-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
5NGJ
 
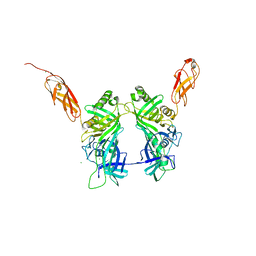 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
5NG1
 
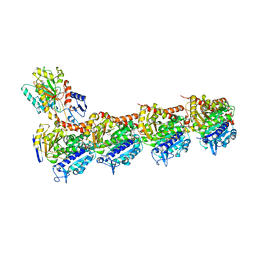 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
2GJS
 
 | | The crystal structure of human RRAD in complex with GDP | | Descriptor: | GTP-binding protein RAD, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Turnbull, A.P, Yang, X, Soundararajan, M, Schoch, G, Debreczeni, J, Elkins, J.M, Gileadi, C, Berridge, G, Pantic, N, Burgess, N, Smee, C.E.A, Bray, J, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human RRAD in complex with GDP
To be Published
|
|
6GGS
 
 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
5MKC
 
 | | Crystal structure of the RrgA Jo.In complex | | Descriptor: | CALCIUM ION, Cell wall surface anchor family protein (Jo),Cell wall surface anchor family protein (In), NICKEL (II) ION, ... | | Authors: | Bonnet, J, Cartannaz, J, Tourcier, G, Contreras-Martel, C, Kleman, J.P, Fenel, D, Schoehn, G, Morlot, C, Vernet, T, Di Guilmi, A.M. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Autocatalytic association of proteins by covalent bond formation: a Bio Molecular Welding toolbox derived from a bacterial adhesin.
Sci Rep, 7, 2017
|
|
7XXN
 
 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7Y4J
 
 | | HapR_Triple mutant Y76F, L97I, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXS
 
 | | HapR mutant I141V | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XYI
 
 | | HapR Quadruple mutant Y76F, L97I, I141V, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXO
 
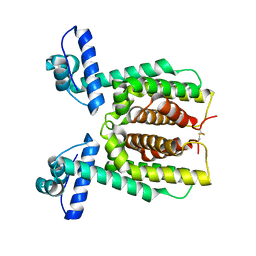 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY5
 
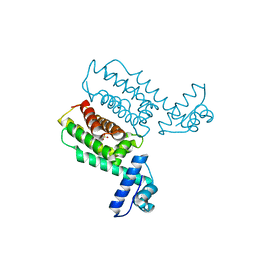 | | HapR_Double Mutant with CHES buffer | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXT
 
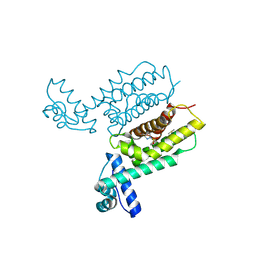 | | HapR Quadruple mutant, bound to IMTVC-212 | | Descriptor: | 1-((5-phenylthiophen-2-yl)sulfonyl)-1H-pyrazole, DIMETHYL SULFOXIDE, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
7OZ4
 
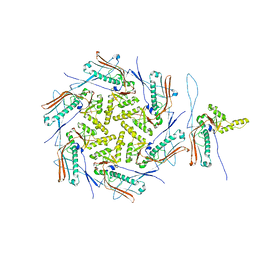 | | Mature capsid of bacteriophage phiRSA1 | | Descriptor: | p2 family phage major capsid protein | | Authors: | Effantin, G, Fujiwara, A, Kawsaki, T, Yamada, T, Schoehn, G. | | Deposit date: | 2021-06-25 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High Resolution Structure of the Mature Capsid of Ralstonia solanacearum Bacteriophage phi RSA1 by Cryo-Electron Microscopy.
Int J Mol Sci, 22, 2021
|
|
2G3Y
 
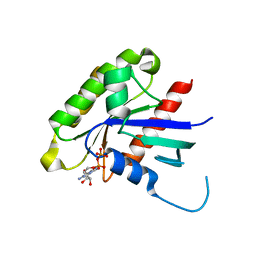 | | Crystal structure of the human small GTPase GEM | | Descriptor: | GTP-binding protein GEM, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ugochukwu, E, Soundararajan, M, Elkins, J, Gileadi, C, Schoch, G, Sobott, F, Fedorov, O, Bray, J, Pantic, N, Berridge, G, Burgess, N, Lee, W.H, Turnbull, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human small GTPase GEM
To be Published
|
|
2FCF
 
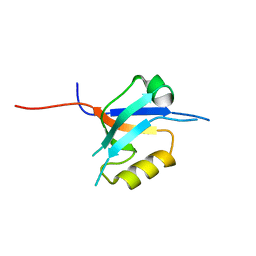 | | The crystal structure of the 7th PDZ domain of MPDZ (MUPP-1) | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2FNE
 
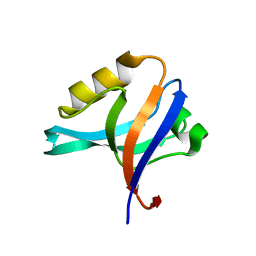 | | The crystal structure of the 13th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
3ZN8
 
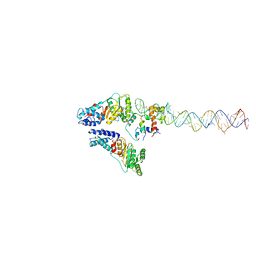 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
2IWO
 
 | | 12th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Yang, X, Gileadi, C, Schoch, G, Johansson, C, Savitsky, P, Berridge, G, Smee, C.E.A, Turnbull, A, Pike, A, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extensions.
Protein Sci., 16, 2007
|
|
2IWP
 
 | | 12th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Yang, X, Gileadi, C, Schoch, G, Johansson, C, Savitsky, P, Berridge, G, Smee, C.E.A, Turnbull, A, Pike, A, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extension.
Protein Sci., 16, 2007
|
|
7ZAY
 
 | | Human heparan sulfate polymerase complex EXT1-EXT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-1, Exostosin-2, ... | | Authors: | Leisico, F, Omeiri, J, Hons, M, Schoehn, G, Lortat-Jacob, H, Wild, R. | | Deposit date: | 2022-03-23 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human heparan sulfate polymerase complex EXT1-EXT2.
Nat Commun, 13, 2022
|
|
4WG0
 
 | |
