6IGU
 
 | | Crystal structure of the hydrolytic antibody Fab 9C10 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IMMUNOGLOBULIN 9C10 H CHAIN, IMMUNOGLOBULIN 9C10 L CHAIN, ... | | Authors: | Yamaguchi, A, Tada, T, Tsuchiya, Y, Tsumuraya, T, Fujii, I. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the complex of the hydrolytic antibody Fab 9C10 and a transition-state analog
To Be Published
|
|
3AP2
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
7WCE
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with (2S)-2-(tert-Butoxy)-2-(10-fluoro-2-(2-hydroxy-4-methylphenyl)-1,4-dimethyl-5-(methylsulfonyl)-5,6-dihydrophenanthridin-3-yl)acetic acid | | Descriptor: | (2S)-2-[10-fluoranyl-1,4-dimethyl-2-(4-methyl-2-oxidanyl-phenyl)-5-methylsulfonyl-6H-phenanthridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, GLYCEROL, Integrase catalytic, ... | | Authors: | Taoda, Y, Sekiguchi, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of tricyclic HIV-1 integrase-LEDGF/p75 allosteric inhibitors by intramolecular direct arylation reaction.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
7WAF
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAC
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | | Descriptor: | Cyanophycin synthase | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAD
 
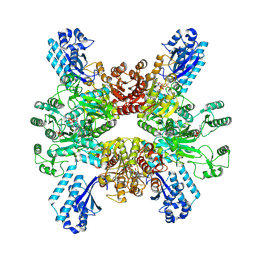 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS | | Descriptor: | Cyanophycin synthase, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAE
 
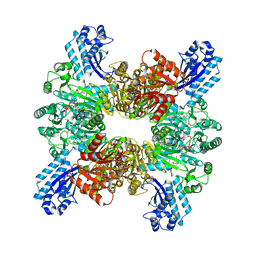 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
3WBD
 
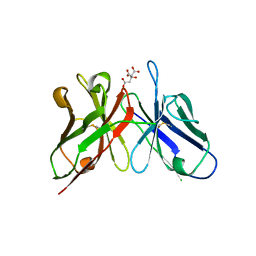 | | Crystal structure of anti-polysialic acid antibody single chain Fv fragment (mAb735) complexed with octasialic acid | | Descriptor: | CITRATE ANION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, single chain Fv fragment of mAb735 | | Authors: | Nagae, M, Ikeda, A, Hanashima, S, Kitajima, K, Sato, C, Yamaguchi, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: insight into the binding preference for polysialic acid.
J.Biol.Chem., 288, 2013
|
|
3WOG
 
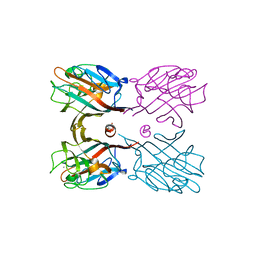 | | Crystal structure plant lectin in complex with ligand | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-12-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3AP3
 
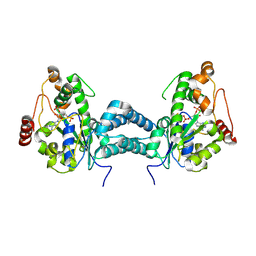 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
3VYK
 
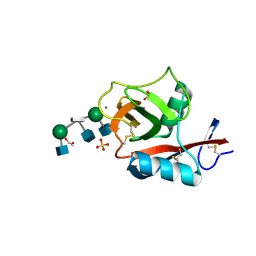 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 in complex with N-glycan | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl alpha-D-mannopyranoside, C-type lectin domain family 4, ... | | Authors: | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
3WCR
 
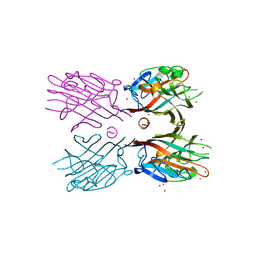 | | Crystal structure of plant lectin (ligand-free form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Erythroagglutinin | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3WCS
 
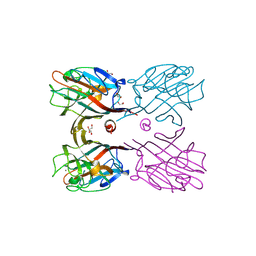 | | Crystal structure of plant lectin (ligand-bound form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
7WCQ
 
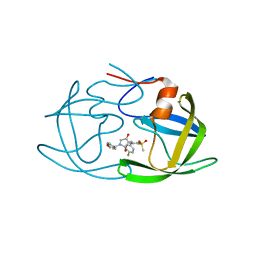 | | Crystal structure of HIV-1 protease in complex with lactam derivative 1 | | Descriptor: | (3R,4R)-3-[(4-fluorophenyl)methyl]-1-[(4-methoxyphenyl)methyl]-3-(4-methylsulfonylphenyl)-4-oxidanyl-pyrrolidin-2-one, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakasa-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
7WBS
 
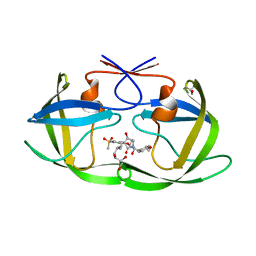 | | Crystal structure of HIV-1 protease in complex with lactam derivative 2 | | Descriptor: | (3~{R},4~{R})-1-[(4-methoxyphenyl)methyl]-3-(3-methylbutyl)-3-[4-methylsulfonyl-2-[(2~{S})-1-oxidanylpropan-2-yl]oxy-phenyl]-4-oxidanyl-pyrrolidin-2-one, GLYCEROL, Protease | | Authors: | Kojima, E, Iimuro, A, Nakajima, M, Kinuta, H, Asada, N, Sako, Y, Nakata, Z, Uemura, K, Arita, S, Miki, S, Wakabayashi-Morimoto, C, Tachibana, Y, Fumoto, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pocket-to-Lead: Structure-Based De Novo Design of Novel Non-peptidic HIV-1 Protease Inhibitors Using the Ligand Binding Pocket as a Template.
J.Med.Chem., 65, 2022
|
|
8J83
 
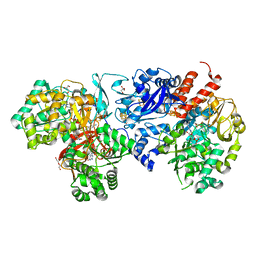 | | Crystal structure of formate dehydrogenase from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kobayashi, A, Taketa, M, Sowa, K, Kano, K, Higuchi, Y, Ogata, H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function relationship of formate dehydrogenases: an overview of recent progress.
Iucrj, 10, 2023
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
5B1O
 
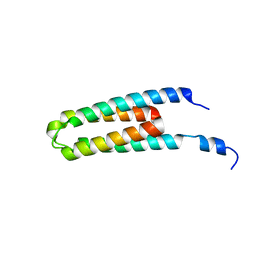 | | DHp domain structure of EnvZ P248A mutant | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
4E1O
 
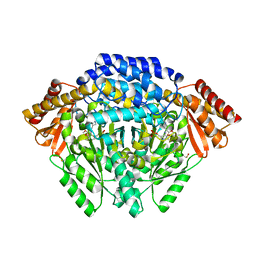 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | Descriptor: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | Deposit date: | 2012-03-06 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
1WQ2
 
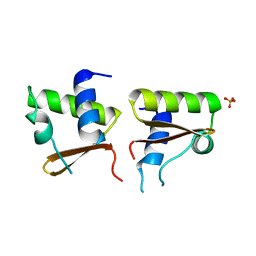 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
7DCZ
 
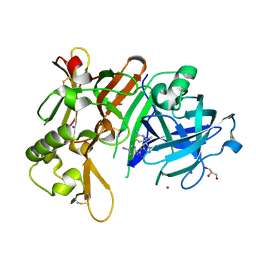 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
5XYO
 
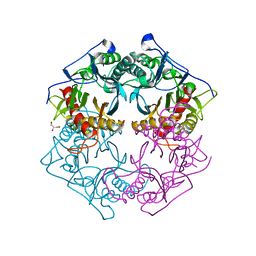 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYP
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122R mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5B1N
 
 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
