4WR4
 
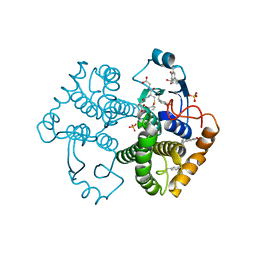 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
7PN0
 
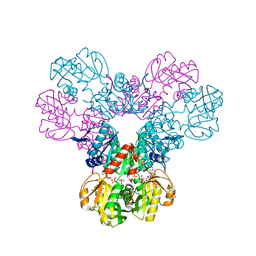 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Timofeev, V.I, Abramchik, Y.A, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-09-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group
To Be Published
|
|
7VTJ
 
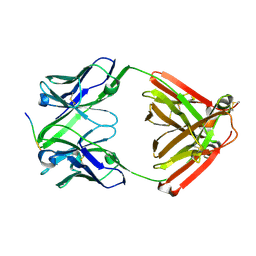 | | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain. | | Descriptor: | Heavy chain of Fab, Light chain of Fab, VQIIYK peptide | | Authors: | Tsuchida, T, Fukuhara, N, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, Y, Ishida, T, Tomoo, K. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cross-reaction complex structure with VQIIYK peptide and tau antibody's Fab domain.
To Be Published
|
|
7WGQ
 
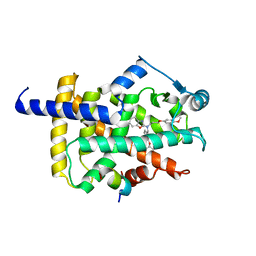 | | X-ray structure of human PPAR gamma ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGO
 
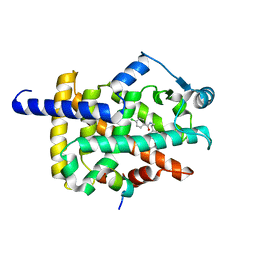 | | X-ray structure of human PPAR gamma ligand binding domain-bezafibrate co-rystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGN
 
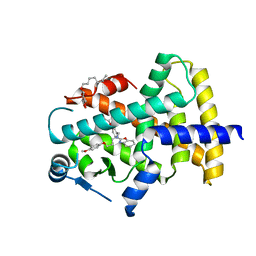 | | X-ray structure of human PPAR delta ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGP
 
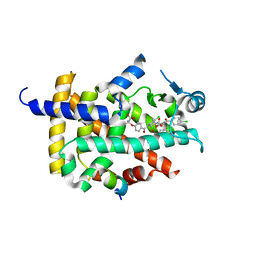 | | X-ray structure of human PPAR gamma ligand binding domain-fenofibric acid co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGL
 
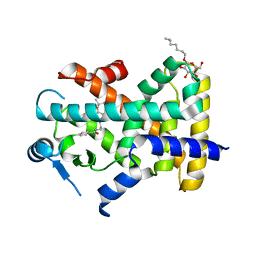 | | X-ray structure of human PPAR delta ligand binding domain-bezafibrate co-crystals obtained by co-crystallization | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
1IYF
 
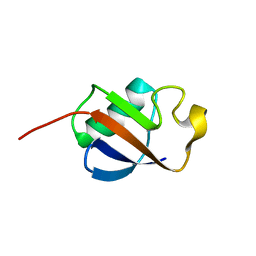 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
5KJS
 
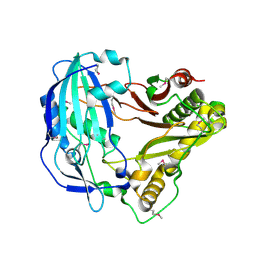 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJV
 
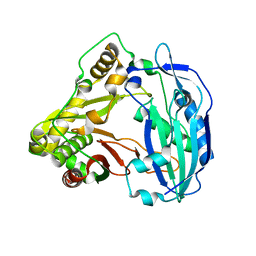 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJT
 
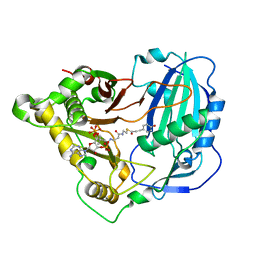 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
8FD4
 
 | |
5KJW
 
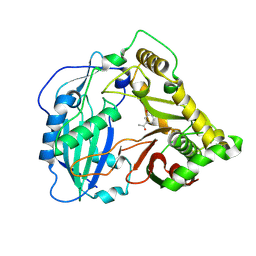 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
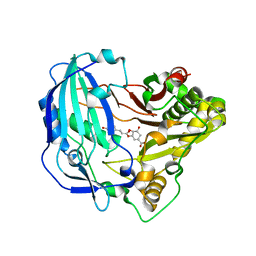 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
6A8U
 
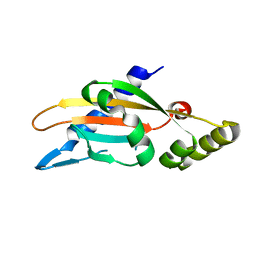 | | PhoQ sensor domain (wild type): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
7E5O
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
6A8V
 
 | | PhoQ sensor domain (D179R mutant): analysis of internal cavity | | Descriptor: | Sensor protein PhoQ | | Authors: | Yoshitani, K, Ishii, E, Taniguchi, K, Sugimoto, H, Shiro, Y, Mori, H, Akiyama, Y, Kato, A, Utsumi, R, Eguchi, Y. | | Deposit date: | 2018-07-10 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control.
Biosci. Biotechnol. Biochem., 83, 2019
|
|
7PZO
 
 | | mite allergen Der p 3 from Dermatophagoides pteronyssinus | | Descriptor: | SULFATE ION, mite allergen Der p 3 | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Mikheeva, O.O, Kostromina, M.A, Lykoshin, D.D, Zayats, E.A, Zavriev, S.K, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
1INL
 
 | | Crystal Structure of Spermidine Synthase from Thermotoga Maritima | | Descriptor: | Spermidine synthase | | Authors: | Korolev, S, Skarina, T, Ikeguchi, Y, Pegg, A.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
7WAF
 
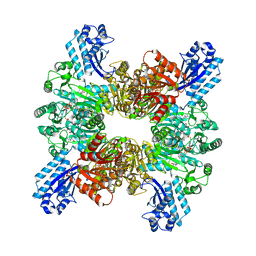 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAE
 
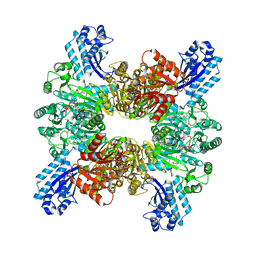 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
8JBQ
 
 | |
1JQ3
 
 | | Crystal Structure of Spermidine Synthase in Complex with Transition State Analogue AdoDATO | | Descriptor: | S-ADENOSYL-1,8-DIAMINO-3-THIOOCTANE, Spermidine synthase | | Authors: | Korolev, S, Ikeguchi, Y, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Pegg, A.E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
3EOQ
 
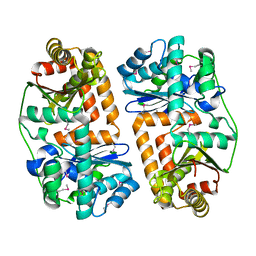 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | Descriptor: | Putative zinc protease | | Authors: | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-09-29 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
