6CHT
 
 | | HNF4alpha in complex with the corepressor EBP1 fragment | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID, Proliferation-associated protein 2G4 | | Authors: | Chi, Y.I, Singh, P, Lee, I.K. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | ErbB3-binding protein 1 (EBP1) represses HNF4 alpha-mediated transcription and insulin secretion in pancreatic beta-cells.
J.Biol.Chem., 294, 2019
|
|
1QVB
 
 | | CRYSTAL STRUCTURE OF THE BETA-GLYCOSIDASE FROM THE HYPERTHERMOPHILE THERMOSPHAERA AGGREGANS | | Descriptor: | BETA-GLYCOSIDASE | | Authors: | Chi, Y.-I, Martinez-Cruz, L.A, Swanson, R.V, Robertson, D.E, Kim, S.-H. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability.
FEBS Lett., 445, 1999
|
|
2QUW
 
 | |
2QUS
 
 | |
4NQZ
 
 | |
4NR0
 
 | |
1CK1
 
 | |
4ZMS
 
 | | Structure of the full-length response regulator spr1814 in complex with a phosphate analogue and B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-diyl dihydroperoxide, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Chi, Y.M, Park, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the full-length response regulator spr1814 in complex with a phosphate analogue reveals a novel conformational plasticity of the linker region
Biochem.Biophys.Res.Commun., 473, 2016
|
|
4ZMR
 
 | |
1CFM
 
 | | CYTOCHROME F FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | CYTOCHROME F, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chi, Y.I, Huang, L.S, Zhang, Z, Fernandez-Velasco, J.G, Malkin, R, Berry, E.A. | | Deposit date: | 1998-09-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of a truncated form of cytochrome f from chlamydomonas reinhardtii.
Biochemistry, 39, 2000
|
|
4QIU
 
 | |
1IC8
 
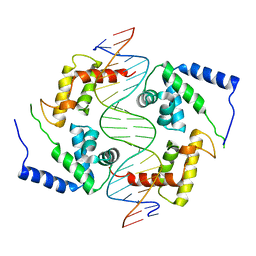 | | HEPATOCYTE NUCLEAR FACTOR 1A BOUND TO DNA : MODY3 GENE PRODUCT | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*G)-3', HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Chi, Y.-I, Frantz, J.D, Oh, B.-C, Hansen, L, Dhe-Paganon, S, Shoelson, S.E. | | Deposit date: | 2001-03-30 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Diabetes mutations delineate an
atypical POU domains in HNF1-Alpha
Mol.Cell, 10, 2002
|
|
4QIT
 
 | |
4QIS
 
 | |
7LVN
 
 | |
2YXV
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2YZ3
 
 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
2FMY
 
 | |
6K31
 
 | |
4UBQ
 
 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
6KBR
 
 | |
6KRN
 
 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, Mating factor alpha,GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6KRU
 
 | | Crystal structure of mouse IgG2b Fc | | Descriptor: | Ig gamma-2B chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
6LFU
 
 | | Poa1p F152A mutant in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.123 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6OVJ
 
 | |
