4D57
 
 | | Understanding bi-specificity of A-domains | | 分子名称: | ADENOSINE MONOPHOSPHATE, APNA A1, ARGININE, ... | | 著者 | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | 登録日 | 2014-11-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3ALR
 
 | | Crystal structure of Nanos | | 分子名称: | Nanos protein, ZINC ION | | 著者 | Hashimoto, H, Hara, K, Hishiki, A, Kawaguchi, S, Shichijo, N, Nakamura, K, Unzai, S, Tamaru, Y, Shimizu, T, Sato, M. | | 登録日 | 2010-08-06 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of zinc-finger domain of Nanos and its functional implications
Embo Rep., 11, 2010
|
|
4D4H
 
 | | Understanding bi-specificity of A-domains | | 分子名称: | APNAA1, GLYCEROL | | 著者 | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | 登録日 | 2014-10-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.019 Å) | | 主引用文献 | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4I
 
 | | Understanding bi-specificity of A-domains | | 分子名称: | APNAA1, ARGININE, GLYCEROL, ... | | 著者 | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | 登録日 | 2014-10-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | 分子名称: | Collagen-like peptide | | 著者 | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | 登録日 | 2011-07-26 | | 公開日 | 2012-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
4D56
 
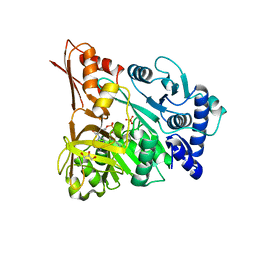 | | Understanding bi-specificity of A-domains | | 分子名称: | ADENOSINE MONOPHOSPHATE, APNAA1, GLYCEROL, ... | | 著者 | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | 登録日 | 2014-11-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2D17
 
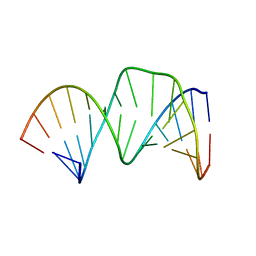 | | Solution RNA structure of stem-bulge-stem region of the HIV-1 dimerization initiation site | | 分子名称: | 5'-R(*CP*GP*GP*CP*AP*AP*GP*AP*GP*GP*CP*GP*AP*CP*CP*C)-3', 5'-R(*GP*GP*GP*UP*CP*GP*GP*CP*UP*UP*GP*CP*UP*G)-3' | | 著者 | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | 登録日 | 2005-08-15 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1A
 
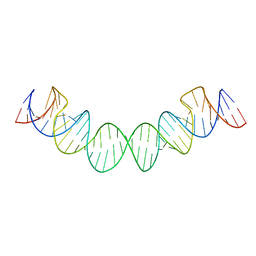 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | 分子名称: | RNA | | 著者 | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | 登録日 | 2005-08-15 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D18
 
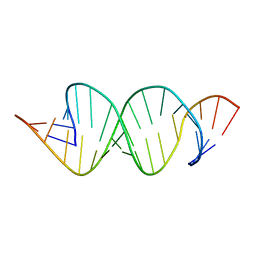 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the extended-duplex dimer | | 分子名称: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | 著者 | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | 登録日 | 2005-08-15 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2DDS
 
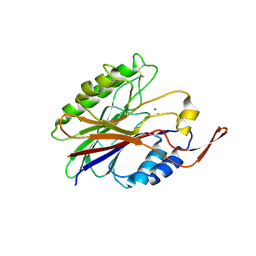 | | Crystal structure of sphingomyelinase from Bacillus cereus with cobalt ion | | 分子名称: | COBALT (II) ION, Sphingomyelin phosphodiesterase | | 著者 | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-02-02 | | 公開日 | 2006-05-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
4A0S
 
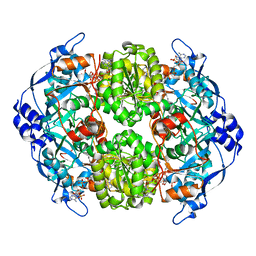 | | STRUCTURE OF THE 2-OCTENOYL-COA CARBOXYLASE REDUCTASE CINF IN COMPLEX WITH NADP AND 2-OCTENOYL-COA | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A, OCTENOYL-COA REDUCTASE/CARBOXYLASE | | 著者 | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | 登録日 | 2011-09-12 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unusual carbon fixation gives rise to diverse polyketide extender units.
Nat. Chem. Biol., 8, 2011
|
|
2D1R
 
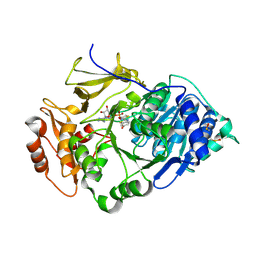 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | 分子名称: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | 著者 | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-08-31 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1Q
 
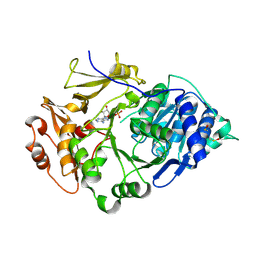 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | 著者 | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-08-31 | | 公開日 | 2006-03-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
3ZFR
 
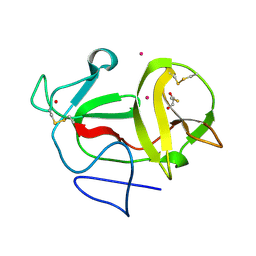 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | 分子名称: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFT
 
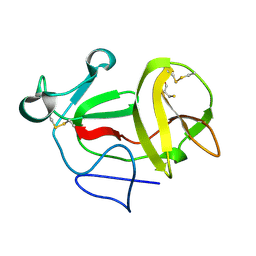 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | 分子名称: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFU
 
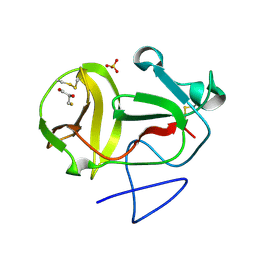 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | 分子名称: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
2D1B
 
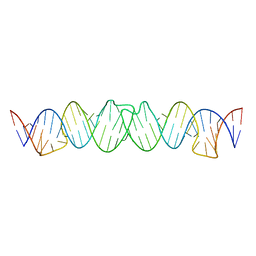 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | 分子名称: | RNA | | 著者 | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | 登録日 | 2005-08-15 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
4A10
 
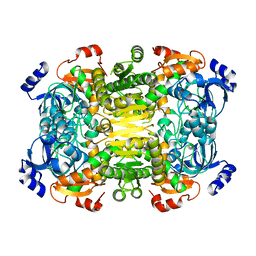 | | Apo-structure of 2-octenoyl-CoA carboxylase reductase CinF from streptomyces sp. | | 分子名称: | OCTENOYL-COA REDUCTASE/CARBOXYLASE | | 著者 | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | 登録日 | 2011-09-13 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Unusual Carbon Fixation Giving Rise to Diverse Polyketide Extender Units
Nat.Chem.Biol., 8, 2011
|
|
4B3A
 
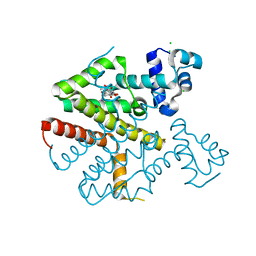 | | Tetracycline repressor class D mutant H100A in complex with tetracycline | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, TETRACYCLINE, ... | | 著者 | Eltschkner, S, Schindler, S, Palm, G.J, Schneider, J, Hinrichs, W. | | 登録日 | 2012-07-23 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Thermodynamics, cooperativity and stability of the tetracycline repressor (TetR) upon tetracycline binding.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
7KXS
 
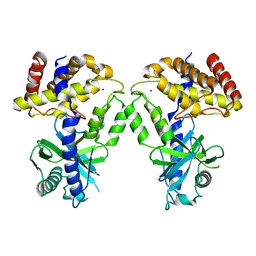 | | Computational design of constitutively active cGAS | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | 登録日 | 2020-12-04 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3ZFQ
 
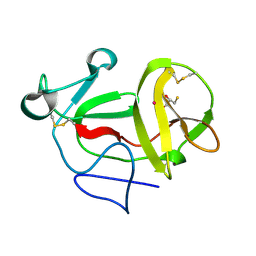 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | 分子名称: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFN
 
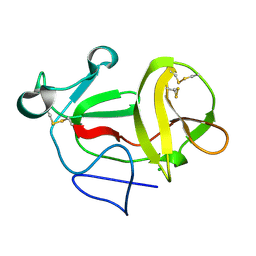 | | Crystal structure of product-like, processed N-terminal protease Npro | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
2D19
 
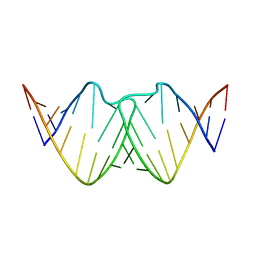 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the kissing-loop dimer | | 分子名称: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | 著者 | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | 登録日 | 2005-08-15 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2CZ4
 
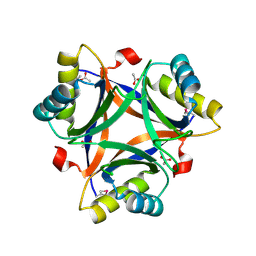 | | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8 | | 分子名称: | ACETATE ION, CHLORIDE ION, hypothetical protein TTHA0516 | | 著者 | Arai, R, Fusatomi, E, Kukimoto-Niino, M, Kawaguchi, S, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-10 | | 公開日 | 2006-01-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structure of a putative PII-like signaling protein (TTHA0516) from Thermus thermophilus HB8
To be Published
|
|
