1UBU
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBM
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBT
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBO
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UBL
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
5BJ3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 1 | | Descriptor: | PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1IO8
 
 | | Thermophilic cytochrome P450 (CYP119) from sulfolobus solfataricus: High resolution structural origin of its thermostability and functional properties | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties
J.Inorg.Biochem., 91, 2002
|
|
8CS7
 
 | | [CCG/CCG] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*CP*CP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.67 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS6
 
 | | [AGG/CTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*CP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS2
 
 | | [(1AP)G/TC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*(1AP)P*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8H8X
 
 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
8CS8
 
 | | [High G:C, High Vapor Diffusion] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*GP*CP*CP*TP*GP*CP*GP*CP*GP*GP*AP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS3
 
 | | [CAG/TCG] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.14 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS4
 
 | | [AGC/GTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.03 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS5
 
 | | [GGA/CTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.52 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5JQP
 
 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | Authors: | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
8CYM
 
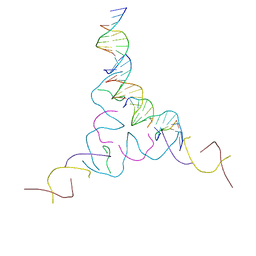 | | [2T7+9bp Linker] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle with 9 bp Sticky-End Linker | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*CP*GP*AP*GP*T)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.76 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DAG
 
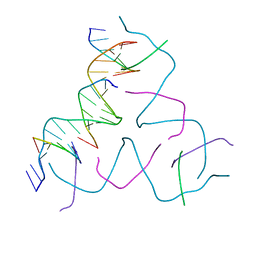 | | [8 bp center] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*TP*GP*TP*GP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.16 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DAH
 
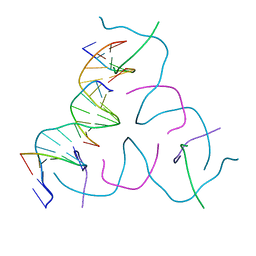 | | [20 bp edge] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.47 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DCJ
 
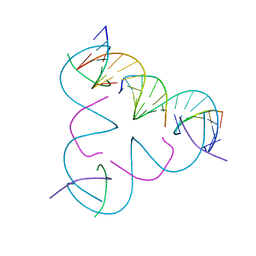 | | [A:T] Self-Assembled 3D DNA Rhombohedral Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8DAP
 
 | | [GA/TC] Self-Assembled 3D DNA Tensegrity Triangle with 24 bp Arm Length forming a Trigonal Hexagon | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*AP*CP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*AP*GP*GP*C)-3'), DNA (5'-D(P*TP*AP*CP*AP*CP*CP*GP*TP*AP*CP*AP*CP*CP*GP*TP*AP*CP*AP*CP*CP*G)-3') | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.47 Å) | | Cite: | Highly Symmetric, Self-Assembling 3D DNA Crystals with Cubic and Trigonal Lattices.
Small, 19, 2023
|
|
8CYN
 
 | | [2T7+20bp Linker] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle with 20 bp Sticky-End Linker | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.45 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8D31
 
 | | [AG/TC] Self-Assembled 3D DNA Cubic Tensegrity Triangle with 24 bp Arm Length | | Descriptor: | DNA (5'-D(*AP*GP*GP*CP*CP*TP*AP*CP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*AP*GP*GP*CP*CP*TP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-31 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.45 Å) | | Cite: | Highly Symmetric, Self-Assembling 3D DNA Crystals with Cubic and Trigonal Lattices.
Small, 19, 2023
|
|
4GH8
 
 | | Crystal structure of a 'humanized' E. coli dihydrofolate reductase | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | French, J.B, Liu, C.T, Hanoian, P, Pringle, T.H, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
