8UQR
 
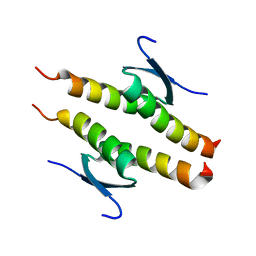 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
7NCS
 
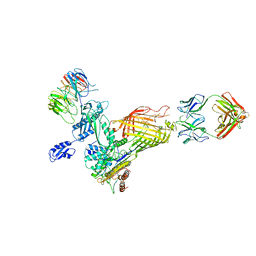 | |
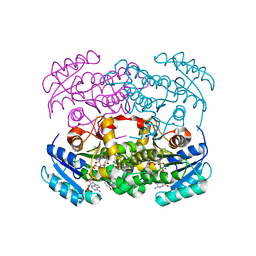
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
6YUR
 
 | | Crystal structure of S. aureus FabI inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weinrich, J.D, Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
6YUU
 
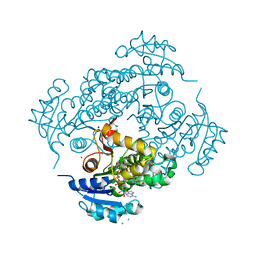 | | Crystal structure of M. tuberculosis InhA inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Weinrich, J.D, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
8TEX
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
8TEY
 
 | | Avian Adeno-associated virus - empty capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Hsi, J, Mietzsch, M, Chipman, P, Afione, S, Zeher, A, Huang, R, Chiorini, J, McKenna, R. | | Deposit date: | 2023-07-07 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and antigenic characterization of the avian adeno-associated virus capsid.
J.Virol., 97, 2023
|
|
6VC6
 
 | | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+ | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-phospho-alpha-glucosidase, GLYCEROL, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2019-12-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Mn2+
To Be Published
|
|
5M5B
 
 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
4K71
 
 | | Crystal structure of a high affinity Human Serum Albumin variant bound to the Neonatal Fc Receptor | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, SULFATE ION, ... | | Authors: | Schmidt, M.M, Townson, S.A, Andreucci, A, Dombrowski, C, Erbe, D.V, King, B, Kovalchin, J.T, Masci, A, Murillo, A, Schirmer, E.B, Furfine, E.S, Barnes, T.M. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-23 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface.
Structure, 21, 2013
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | Descriptor: | Hoc-like protein ORF90, Major structural protein ORF12 | | Authors: | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-02-27 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
5LI0
 
 | | 70S ribosome from Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Bochler, A, Grosse, F, Myasnikov, A, Menetret, J.F, Chicher, J, Marzi, S, Romby, P, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2016-07-13 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the 70S ribosome from human pathogen Staphylococcus aureus.
Nucleic Acids Res., 44, 2016
|
|
2JMI
 
 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
5LWR
 
 | | Endothiapepsin in complex with a derivative of fragment 177 | | Descriptor: | 1,2-ETHANEDIOL, 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
7Q9X
 
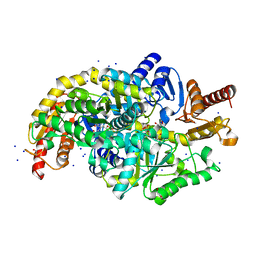 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
7Q9Z
 
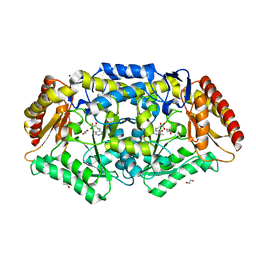 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
2JMJ
 
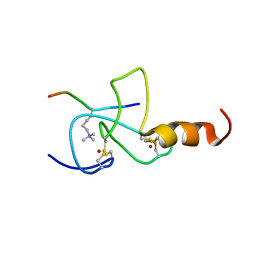 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | Descriptor: | Histone H3, Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
7QW3
 
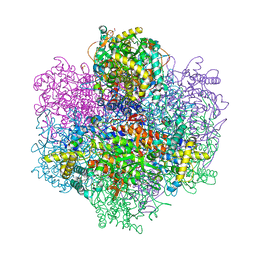 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with Br ion. | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QVW
 
 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QWI
 
 | | Vanadate complex of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
