5V4L
 
 | |
7O2Z
 
 | | Crystal structure of the anti-PAS Fab 2.2 in complex with its epitope peptide | | 分子名称: | CHLORIDE ION, P/A#1 epitope peptide, anti-PAS Fab 2.2 chimeric heavy chain, ... | | 著者 | Schilz, J, Schiefner, A, Skerra, A. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
8I52
 
 | |
4LOX
 
 | |
4LQ0
 
 | |
1IUF
 
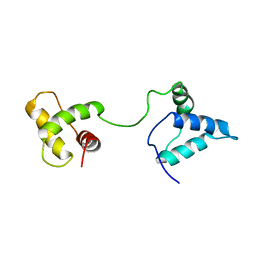 | | LOW RESOLUTION SOLUTION STRUCTURE OF THE TWO DNA-BINDING DOMAINS IN Schizosaccharomyces pombe ABP1 PROTEIN | | 分子名称: | centromere abp1 protein | | 著者 | Kikuchi, J, Iwahara, J, Kigawa, T, Murakami, Y, Okazaki, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-04 | | 公開日 | 2002-06-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure determination of the two DNA-binding domains in the Schizosaccharomyces pombe Abp1 protein by a combination of dipolar coupling and diffusion anisotropy restraints.
J.Biomol.NMR, 22, 2002
|
|
5AXO
 
 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Metallo-beta-lactamase, ... | | 著者 | Wachino, J, Arakawa, Y. | | 登録日 | 2015-07-31 | | 公開日 | 2016-05-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Meropenem
To Be Published
|
|
5V33
 
 | | R. sphaeroides photosythetic reaction center mutant - Residue L223, Ser to Trp - Room Temperature Structure Solved on X-ray Transparent Microfluidic Chip | | 分子名称: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | 著者 | Schieferstein, J.M, Pawate, A.S, Sun, C, Wan, F, Broecker, J, Ernst, O.P, Gennis, R.B, Kenis, P.J.A. | | 登録日 | 2017-03-06 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.487 Å) | | 主引用文献 | X-ray transparent microfluidic chips for high-throughput screening and optimization of in meso membrane protein crystallization.
Biomicrofluidics, 11, 2017
|
|
5AYA
 
 | |
5LWS
 
 | | Endothiapepsin in complex with fragment 177 and a derivative thereof | | 分子名称: | 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, 4-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazine, ACETATE ION, ... | | 著者 | Schiebel, J, Heine, A, Klebe, G. | | 登録日 | 2016-09-19 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7AWT
 
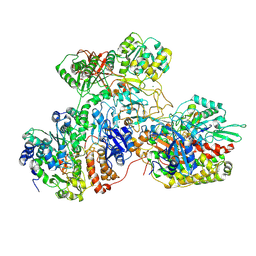 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
5LWU
 
 | | Structure resulting from an endothiapepsin crystal soaked with a dimeric derivative of fragment 177 | | 分子名称: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | 著者 | Schiebel, J, Heine, A, Klebe, G. | | 登録日 | 2016-09-19 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.109 Å) | | 主引用文献 | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LWT
 
 | | Endothiapepsin in complex with a methoxylated derivative of fragment 177 | | 分子名称: | 4-methoxy-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Schiebel, J, Heine, A, Klebe, G. | | 登録日 | 2016-09-19 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.069 Å) | | 主引用文献 | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3JB5
 
 | |
3TM1
 
 | |
1N6U
 
 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | 分子名称: | Interferon-alpha/beta receptor beta chain | | 著者 | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | 登録日 | 2002-11-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
7B0M
 
 | | Sugar transaminase from a metagenome collected from troll oil field production water | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar aminotransferase, ... | | 著者 | Littlechild, J.A, De Rose, S.A, Isupov, M.N, Sayer, C, Karki, S. | | 登録日 | 2020-11-20 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Sugar transaminases from hot environments
To Be Published
|
|
5MNQ
 
 | | Cationic trypsin in complex with a derivative of N-amidinopiperidine | | 分子名称: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(1-carbamimidoylpiperidin-4-yl)methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | 著者 | Schiebel, J, Ngo, K, Heine, A, Klebe, G. | | 登録日 | 2016-12-13 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.337 Å) | | 主引用文献 | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
3TM2
 
 | | Crystal structure of mature ThnT with a covalently bound product mimic | | 分子名称: | (2R)-N-(4-chloro-3-oxobutyl)-2,4-dihydroxy-3,3-dimethylbutanamide, cysteine transferase | | 著者 | Schildbach, J.F, Wright, N.T, Buller, A.R. | | 登録日 | 2011-08-30 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Autoproteolytic Activation of ThnT Results in Structural Reorganization Necessary for Substrate Binding and Catalysis.
J.Mol.Biol., 422, 2012
|
|
6GNI
 
 | |
1N6V
 
 | | Average structure of the interferon-binding ectodomain of the human type I interferon receptor | | 分子名称: | Interferon-alpha/beta receptor beta chain | | 著者 | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | 登録日 | 2002-11-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
6A6P
 
 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316 | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside, ... | | 著者 | Chin, J.W, Cho, S.J, Song, J.Y, Ha, J.H. | | 登録日 | 2018-06-29 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd)LBD in Complex with DN003316
To Be Published
|
|
1AW0
 
 | |
5DR3
 
 | | Endothiapepsin in complex with fragment 333 | | 分子名称: | 1,2-ETHANEDIOL, 4-propan-2-ylsulfanyl-1-propyl-6,7-dihydro-5~{H}-cyclopenta[d]pyrimidin-2-one, ACETATE ION, ... | | 著者 | Schiebel, J, Heine, A, Klebe, G. | | 登録日 | 2015-09-15 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
7O33
 
 | |
