3R1G
 
 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
4GUY
 
 | | Human MMP12 catalytic domain in complex with*N*-Hydroxy-2-(2-(4-methoxyphenyl)ethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-{[2-(4-methoxyphenyl)ethyl]sulfonyl}glycinamide, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C, Massaro, A, Mordini, A, Mori, M. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of free energy of solvation to ligand affinity in new potent MMPs inhibitors.
To be Published
|
|
1V06
 
 | |
3RTT
 
 | | Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-1-[(2-phenylethyl)sulfonyl]-D-prolinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
2M41
 
 | |
3OE3
 
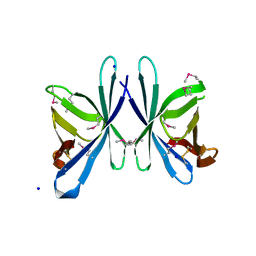 | | Crystal structure of PliC-St, periplasmic lysozyme inhibitor of C-type lysozyme from Salmonella typhimurium | | Descriptor: | Putative periplasmic protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-12 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3RTS
 
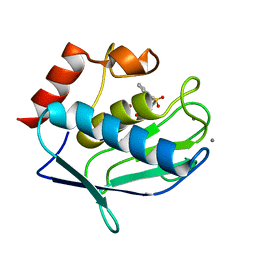 | | Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-[(2-phenylethyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
1KSM
 
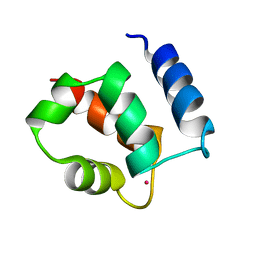 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
3PBB
 
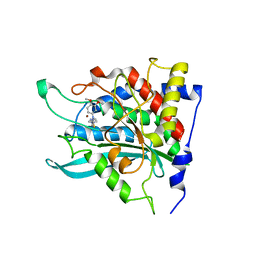 | | Crystal structure of human secretory glutaminyl cyclase in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
2OY2
 
 | | Human MMP-8 in complex with peptide IAG | | Descriptor: | CALCIUM ION, ILE-ALA-GLY peptide, Neutrophil collagenase, ... | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Maletta, M, Yeo, K.J. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the reaction mechanism of matrix metalloproteinases.
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
3PB6
 
 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase at pH 6.5 | | Descriptor: | CACODYLATE ION, Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
2L51
 
 | | Solution structure of calcium bound S100A16 | | Descriptor: | CALCIUM ION, Protein S100-A16 | | Authors: | Babini, E, Bertini, I, Borsi, V, Calderone, V, Hu, X, Luchinat, C, Parigi, G. | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of human S100A16, a low-affinity calcium binder.
J.Biol.Inorg.Chem., 16, 2011
|
|
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | Descriptor: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
1TUW
 
 | | Structural and Functional Analysis of Tetracenomycin F2 Cyclase from Streptomyces glaucescens: A Type-II Polyketide Cyclase | | Descriptor: | SULFATE ION, Tetracenomycin polyketide synthesis protein tcmI | | Authors: | Thompson, T.B, Katayama, K, Watanabe, K, Hutchinson, C.R, Rayment, I. | | Deposit date: | 2004-06-25 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase.
J.Biol.Chem., 279, 2004
|
|
2OY4
 
 | | Uninhibited human MMP-8 | | Descriptor: | CALCIUM ION, Neutrophil collagenase, ZINC ION | | Authors: | Calderone, V, Bertini, I, Fragai, M, Luchinat, C, Maletta, M, Yeo, K.J. | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Snapshots of the reaction mechanism of matrix metalloproteinases.
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
5J8W
 
 | | One minute iron loaded Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
6PIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PIW
 
 | |
1SW8
 
 | | Solution structure of the N-terminal domain of Human N60D calmodulin refined with paramagnetism based strategy | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Bertini, I, Del Bianco, C, Gelis, I, Katsaros, N, Luchinat, C, Parigi, G, Peana, M, Provenzani, A, Zoroddu, M.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally exploring the conformational space sampled by domain reorientation in calmodulin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4IOU
 
 | |
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
4IJO
 
 | | Unraveling hidden allosteric regulatory sites in structurally homologues metalloproteases | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, ZINC ION | | Authors: | Udi, Y, Fragai, M, Grossman, M, Mitternacht, S, Arad-Yellin, R, Calderone, V, Melikian, M, Toccafondi, M, Berezovsky, I.N, Luchinat, C, Sagi, I. | | Deposit date: | 2012-12-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling hidden regulatory sites in structurally homologous metalloproteases
J.Mol.Biol., 425, 2013
|
|
5YEQ
 
 | | The structure of Sac-KARI protein | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Ko, T.P, Chen, C.Y, Lin, K.F, Lin, B.L, Huang, C.H, Chiang, C.H, Horng, J.C, Tsai, M.D. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NADH/NADPH bi-cofactor-utilizing and thermoactive ketol-acid reductoisomerase from Sulfolobus acidocaldarius
Sci Rep, 8, 2018
|
|
