8SUN
 
 | | TMEM16F 1PBC | | Descriptor: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, ... | | Authors: | Wu, H, Feng, S, Cheng, Y. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
7U25
 
 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase W574L mutant in complex with bispyribac-sodium | | Descriptor: | 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
7U1U
 
 | |
7RMI
 
 | | SP6-11 biased agonist bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMG
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMH
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
6O72
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, TC-I 2014-bound state | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
6UTG
 
 | |
3WRE
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
2B6O
 
 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
7MZ5
 
 | |
7MZ6
 
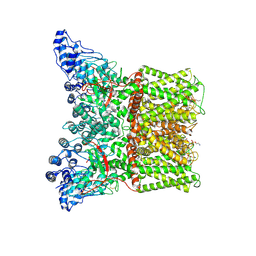 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZB
 
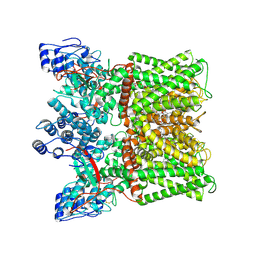 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
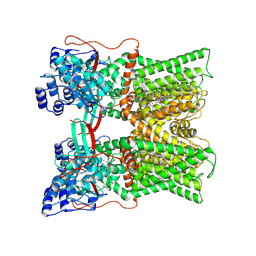 | |
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ9
 
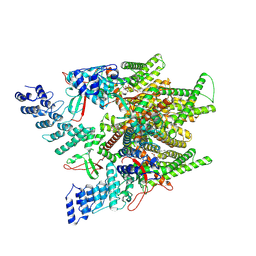 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZD
 
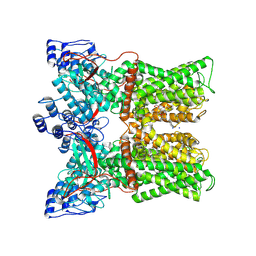 | |
7MZE
 
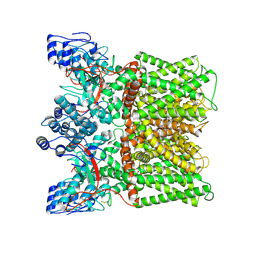 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in opposite pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
2B6P
 
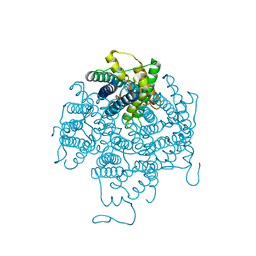 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
1PT3
 
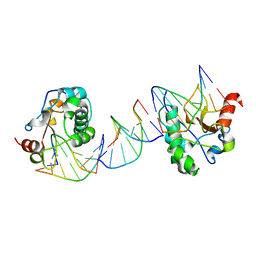 | | Crystal structures of nuclease-ColE7 complexed with octamer DNA | | Descriptor: | 5'-GCGATCGC-3', Colicin E7 | | Authors: | Hsia, K.C, Chak, K.F, Cheng, Y.S, Ku, W.Y, Yuan, H.S. | | Deposit date: | 2003-06-22 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding and degradation by the HNH protein ColE7.
STRUCTURE, 12, 2004
|
|
1Q57
 
 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
