7JV5
 
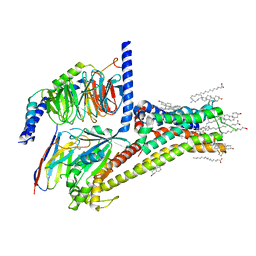 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
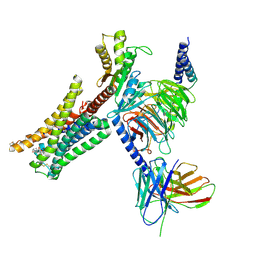 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
2FS4
 
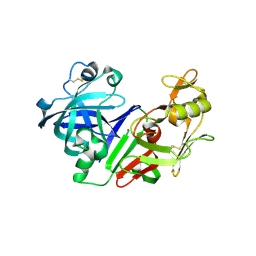 | | Ketopiperazine-Based Renin Inhibitors: Optimization of the C ring | | Descriptor: | (6R)-6-({[1-(3-HYDROXYPROPYL)-1,7-DIHYDROQUINOLIN-7-YL]OXY}METHYL)-1-(4-{3-[(2-METHOXYBENZYL)OXY]PROPOXY}PHENYL)PIPERAZIN-2-ONE, Renin | | Authors: | Holsworth, D.D, Cai, C, Cheng, X.-M, Cody, W.L, Downing, D.M, Erasga, N, Lee, C, Powell, N.A, Edmunds, J.J, Stier, M, Jalaie, M, Zhang, E, McConnell, P, Ryan, M.J, Bryant, J, Li, T, Kasani, A, Hall, E, Subedi, R, Rahim, M, Maiti, S. | | Deposit date: | 2006-01-20 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ketopiperazine-Based Renin Inhibitors: Optimization of the "C" Ring
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
7JVQ
 
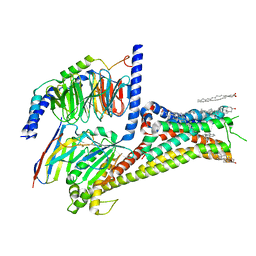 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
3FPD
 
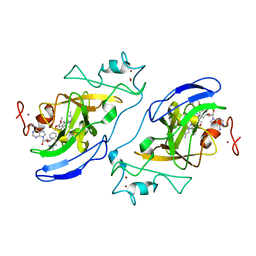 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8F8Y
 
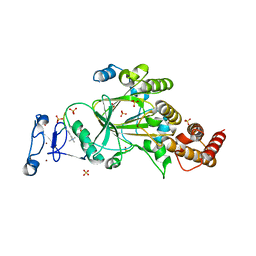 | | PHF2 (PHD+JMJ) in Complex with VRK1 N-Terminal Peptide | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase PHF2, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A complete methyl-lysine binding aromatic cage constructed by two domains of PHF2.
J.Biol.Chem., 299, 2022
|
|
8F8Z
 
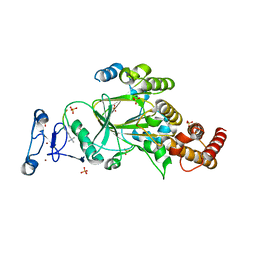 | |
8FS1
 
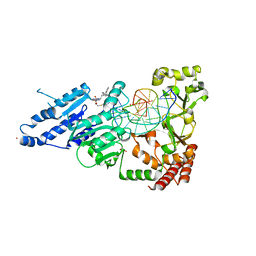 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
1EYU
 
 | |
1F0O
 
 | |
3B7B
 
 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 1) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules
Nat.Struct.Mol.Biol., 15, 2008
|
|
3B95
 
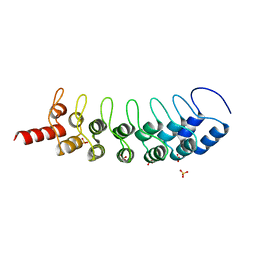 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 2) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, Histone H3 N-terminal Peptide, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3ICW
 
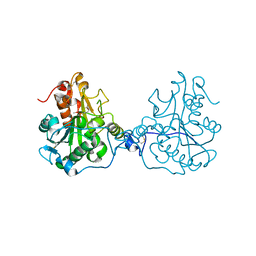 | | Structure of a Circular Permutation on Lipase B from Candida Antartica with Bound Suicide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, PHOSPHATE ION, ... | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S.A, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3ICV
 
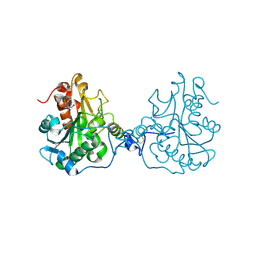 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3EU9
 
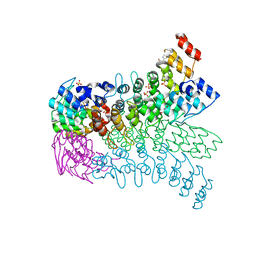 | | The ankyrin repeat domain of Huntingtin interacting protein 14 | | Descriptor: | GLYCEROL, HISTIDINE, Huntingtin-interacting protein 14, ... | | Authors: | Gao, T, Collins, R.E, Horton, J.R, Zhang, R, Zhang, X, Cheng, X. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding.
Proteins, 76, 2009
|
|
6WK1
 
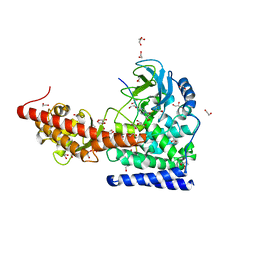 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
5KE6
 
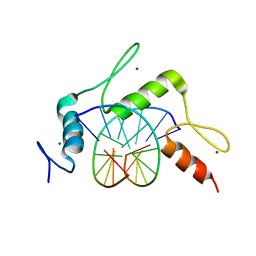 | | mouse Klf4 ZnF1-3 and TpG/CpA sequence DNA complex structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*CP*AP*CP*CP*TP*C)-3'), Krueppel-like factor 4, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Distinctive Klf4 mutants determine preference for DNA methylation status.
Nucleic Acids Res., 44, 2016
|
|
5KEB
 
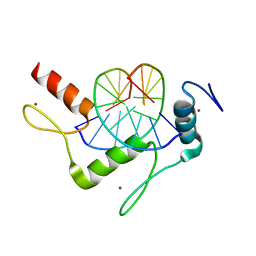 | |
3F8I
 
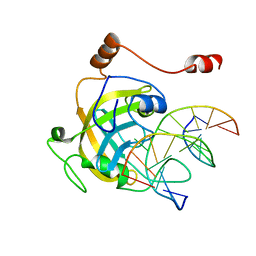 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
5K5I
 
 | |
3FDE
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
6OX5
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
5K5L
 
 | |
