6J21
 
 | | Crystal structure of the human NK1 substance P receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, Substance-P receptor,Endolysin | | Authors: | Chen, S, Lu, M, Zhang, H, Wu, B, Zhao, Q. | | Deposit date: | 2018-12-30 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human substance P receptor binding mode of the antagonist drug aprepitant by NMR and crystallography.
Nat Commun, 10, 2019
|
|
7F5F
 
 | | SARS-CoV-2 ORF8 S84 | | Descriptor: | CALCIUM ION, ORF8 protein | | Authors: | Chen, S, Zhou, Z, Chen, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structures of Bat and Human Coronavirus ORF8 Protein Ig-Like Domain Provide Insights Into the Diversity of Immune Responses.
Front Immunol, 12, 2021
|
|
7CR5
 
 | |
7DE1
 
 | |
3O5Z
 
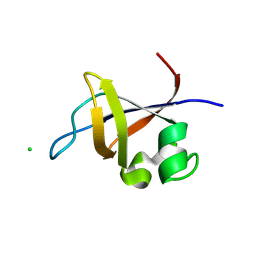 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3P38
 
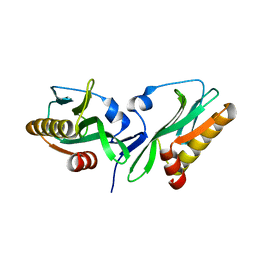 | |
3P31
 
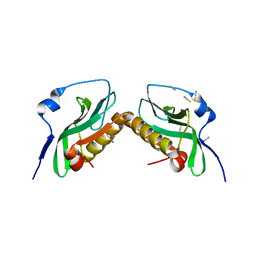 | |
3P39
 
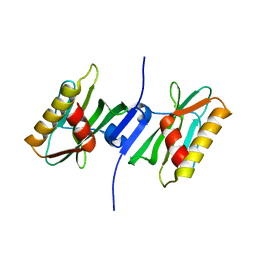 | |
3ZDP
 
 | |
7DHY
 
 | | Arsenic-bound p53 DNA-binding domain mutant G245S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
7DHZ
 
 | | Arsenic-bound p53 DNA-binding domain mutant R249S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
7EDP
 
 | | Crystal structure of AF10-DOT1L complex | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | Authors: | Chen, S, Zhou, Z. | | Deposit date: | 2021-03-16 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characteristics of coiled-coil regions in AF10-DOT1L and AF10-inhibitory peptide complex.
J Leukoc Biol, 110, 2021
|
|
7EKN
 
 | | Crystal structure of AF10-ipep complex | | Descriptor: | Protein AF-10, ipep | | Authors: | Chen, S, Zhou, Z. | | Deposit date: | 2021-04-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural characteristics of coiled-coil regions in AF10-DOT1L and AF10-inhibitory peptide complex.
J Leukoc Biol, 110, 2021
|
|
8K33
 
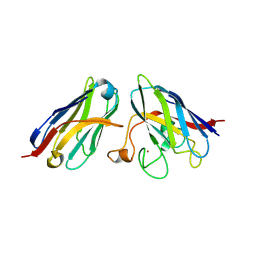 | | SOD1 and Nanobody1 complex | | Descriptor: | NB1, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Cheng, S. | | Deposit date: | 2023-07-14 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | SOD1 and Nanobody1 complex
To Be Published
|
|
8K3A
 
 | | SOD1 and Nanobody2 complex | | Descriptor: | NB2, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Cheng, S. | | Deposit date: | 2023-07-14 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | SOD1 and Nanobody1 complex
To Be Published
|
|
6NRX
 
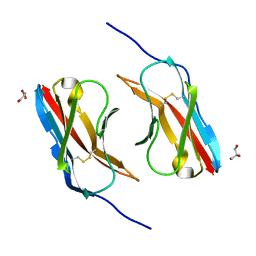 | | Crystal structure of DIP-eta IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein eta, isoform B, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NRQ
 
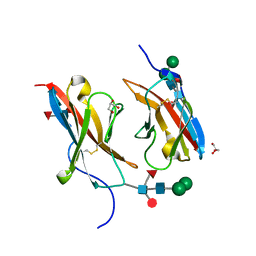 | | Crystal structure of Dpr10 IG1 bound to DIP-alpha IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Defective proboscis extension response 10, isoform A, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NS1
 
 | | Crystal structure of DIP-gamma IG1+IG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein gamma | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NRW
 
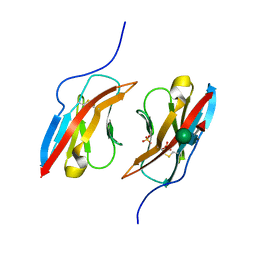 | | Crystal structure of Dpr1 IG1 bound to DIP-eta IG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
6NRR
 
 | | Crystal structure of Dpr11 IG1 bound to DIP-gamma IG+IG2 | | Descriptor: | Defective proboscis extension response 11, isoform B, Dpr-interacting protein gamma, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
1BH6
 
 | | SUBTILISIN DY IN COMPLEX WITH THE SYNTHETIC INHIBITOR N-BENZYLOXYCARBONYL-ALA-PRO-PHE-CHLOROMETHYL KETONE | | Descriptor: | CALCIUM ION, N-BENZYLOXYCARBONYL-ALA-PRO-3-AMINO-4-PHENYL-BUTAN-2-OL, SODIUM ION, ... | | Authors: | Eschenburg, S, Genov, N, Wilson, K.S, Betzel, C. | | Deposit date: | 1998-06-15 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of subtilisin DY, a random mutant of subtilisin Carlsberg.
Eur.J.Biochem., 257, 1998
|
|
1EJD
 
 | | Crystal structure of unliganded mura (type1) | | Descriptor: | CYCLOHEXYLAMMONIUM ION, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
1I6U
 
 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | Authors: | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | Deposit date: | 2001-03-05 | | Release date: | 2001-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
8TKU
 
 | | ZIG-4-INS-6 complex, primitive monoclinic form | | Descriptor: | Probable insulin-like peptide beta-type 5, Zwei Ig domain protein zig-4 | | Authors: | Cheng, S, Baltrusaitis, E, Aziz, Z, Nawrocka, W.I, Ozkan, E. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways.
Biorxiv, 2024
|
|
6PLL
 
 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
